5L17
 
 | |
5L15
 
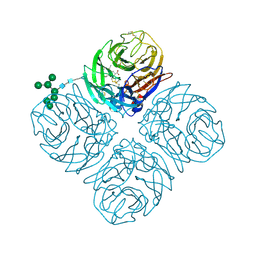 | | The crystal structure of neuraminidase in complex with oseltamivir from A/Shanghai/2/2013 (H7N9) influenza virus | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Drug Susceptibility Evaluation of an Influenza A(H7N9) Virus by Analyzing Recombinant Neuraminidase Proteins.
J. Infect. Dis., 216, 2017
|
|
5L14
 
 | |
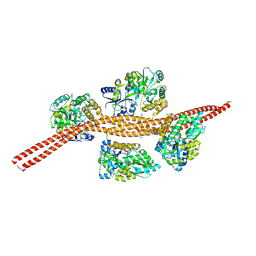
3MQ7
 
 | | Crystal Structure of Ectodomain Mutant of BST-2/Tetherin/CD317 | | Descriptor: | Bone marrow stromal antigen 2, CALCIUM ION | | Authors: | Xiong, Y, Yang, H, Wang, J, Meng, W. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insight into the mechanisms of enveloped virus tethering by tetherin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MQ9
 
 | |
3MQB
 
 | |
3MQC
 
 | |
6E00
 
 | | Structure of a N-Me-p-iodo-D-Phe1,N-Me-D-Gln4,Lys10-teixobactin analogue | | Descriptor: | N-Me-p-iodo-D-Phe1,N-Me-D-Gln4,Lys10-teixobactin analogue, SULFATE ION | | Authors: | Nowick, J.S, Yang, H, Wierzbicki, M. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystallographic Structure of a Teixobactin Derivative Reveals Amyloid-like Assembly.
J. Am. Chem. Soc., 140, 2018
|
|
3PHD
 
 | | Crystal structure of human HDAC6 in complex with ubiquitin | | Descriptor: | Histone deacetylase 6, Polyubiquitin, ZINC ION | | Authors: | Dong, A, Qui, W, Ravichandran, M, Schuetz, A, Loppnau, P, Li, F, Mackenzie, F, Kozieradzki, I, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Aggregates Are Recruited to Aggresome by Histone Deacetylase 6 via Unanchored Ubiquitin C Termini.
J.Biol.Chem., 287, 2012
|
|
4JSX
 
 | | structure of mTORDeltaN-mLST8-Torin2 complex | | Descriptor: | 9-(6-aminopyridin-3-yl)-1-[3-(trifluoromethyl)phenyl]benzo[h][1,6]naphthyridin-2(1H)-one, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
4JSV
 
 | | mTOR kinase structure, mechanism and regulation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase mTOR, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
4JT5
 
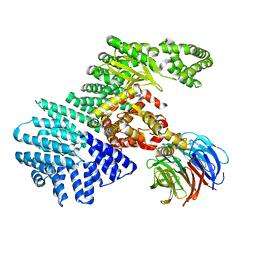 | | mTORdeltaN-mLST8-pp242 complex | | Descriptor: | 2-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-1H-indol-5-ol, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
4JT6
 
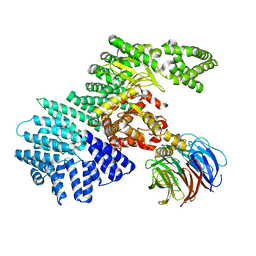 | | structure of mTORDeltaN-mLST8-PI-103 complex | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, mLST8, mTOR | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
4JSP
 
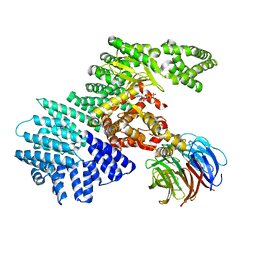 | | structure of mTORDeltaN-mLST8-ATPgammaS-Mg complex | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase mTOR, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
2H2Z
 
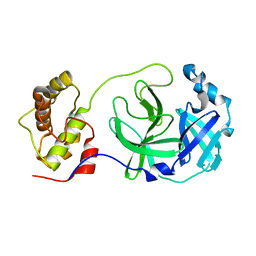 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
4KWM
 
 | | Structure of a/anhui/5/2005 h5 ha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Shore, D.A, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-24 | | Release date: | 2014-06-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and antigenic variation among diverse clade 2 H5N1 viruses.
Plos One, 8, 2013
|
|
4GEZ
 
 | | Structure of a neuraminidase-like protein from A/bat/Guatemala/164/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Carney, P.J, Donis, R.O, Stevens, J. | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4MC5
 
 | | Crystal structure of a subtype H18 hemagglutinin homologue from A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
4MC7
 
 | | Crystal structure of a subtype N11 neuraminidase-like protein of A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
8U78
 
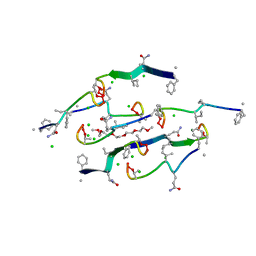 | | Structure of a N-Me-D-Gln4,Lys10-teixobactin analogue | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, N-Methyl-D-Gln4,Lys10-teixobactin | | Authors: | Nowick, J.S, Yang, H, Kreutzer, A.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
J.Org.Chem., 89, 2024
|
|
8JJ5
 
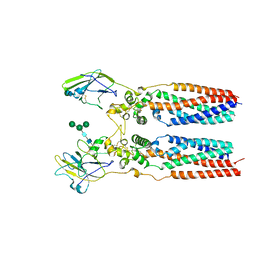 | | Porcine uroplakin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tetraspanin, ... | | Authors: | Oda, T, Yanagisawa, H, Kikkawa, M. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM elucidates the uroplakin complex structure within liquid-crystalline lipids in the porcine urothelial membrane.
Commun Biol, 6, 2023
|
|
8HWN
 
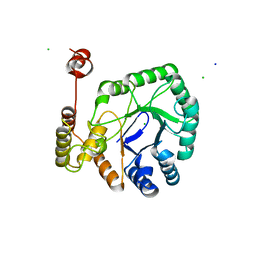 | | aldo-keto reductase DepB | | Descriptor: | CHLORIDE ION, DepB, SODIUM ION | | Authors: | Chen, M, Yang, H, Lu, F. | | Deposit date: | 2022-12-31 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of DepB capable of DON detoxification
To Be Published
|
|
5Y8E
 
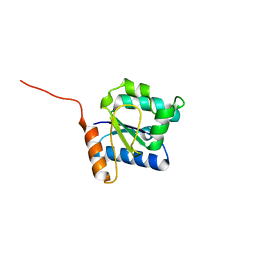 | |
5Y8F
 
 | |
6ANW
 
 | | Crystal structure of anti-CRISPR protein AcrF10 | | Descriptor: | anti-CRISPR protein AcrF10 | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
