7PYK
 
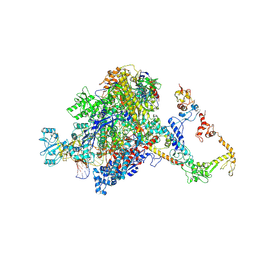 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY5
 
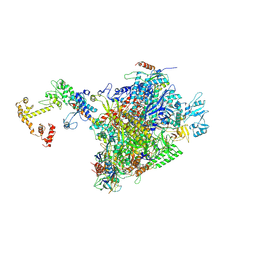 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (the consensus NusA-NusG-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7Q0J
 
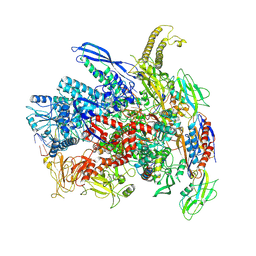 | | RNA polymerase elongation complex in more-swiveled conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY0
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (NusG-EC in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY8
 
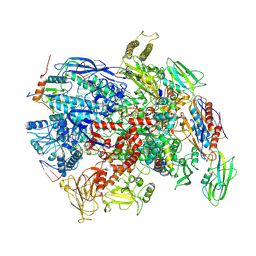 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (NusG-EC in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY3
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (the consensus NusA-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7Q0K
 
 | | RNA polymerase elongation complex in less-swiveled conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY6
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY1
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (the consensus NusG-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PYJ
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
6HG8
 
 | | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Ambrus, A, Szabo, E, Weichsel, A, Bui, D, Wilk, P, Torocsik, B, Weiss, M.S, Montfort, W.R, Jordan, F, Adam-Vizi, V. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase.
To Be Published
|
|
1ML7
 
 | | Crystal structure of nitrophorin 4 complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, nitrophorin 4 | | Authors: | Berry, R.E, Ding, X.D, Weichsel, A, Montfort, W.R, Walker, F.A. | | Deposit date: | 2002-08-30 | | Release date: | 2002-09-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Axial ligand complexes of the Rhodnius nitrophorins: reduction potentials, binding constants, EPR spectra, and structures of the 4-iodopyrazole and imidazole complexes of NP4
J.Biol.Inorg.Chem., 9, 2004
|
|
1PLK
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLJ
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLL
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5A5B
 
 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1N68
 
 | | Copper bound to the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-CL-CU LINKAGE | | Authors: | Roberts, S.A, Wildner, G.F, Grass, G, Weichsel, A, Ambrus, A, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-11-08 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Labile Regulatory Copper Ion Lies Near the T1 Copper Site in the Multicopper Oxidase CueO.
J.Biol.Chem., 278, 2003
|
|
6ZTP
 
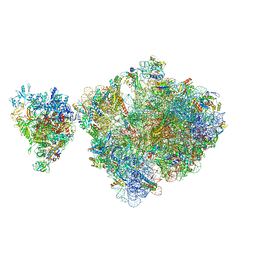 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTN
 
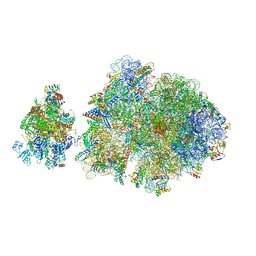 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTJ
 
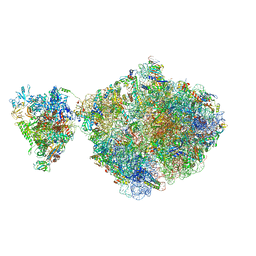 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTO
 
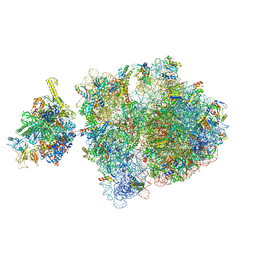 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZU1
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTL
 
 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTM
 
 | | E. coli 70S-RNAP expressome complex in collided state without NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
1F5L
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN-AMILORIDE COMPLEX | | Descriptor: | 3,5-DIAMINO-N-(AMINOIMINOMETHYL)-6-CHLOROPYRAZINECARBOXAMIDE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Zeslawska, E, Schweinitz, A, Karcher, A, Sondermann, P, Sperl, S, Sturzebecher, J, Jacob, U. | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals of the urokinase type plasminogen activator variant beta(c)-uPAin complex with small molecule inhibitors open the way towards structure-based drug design.
J.Mol.Biol., 301, 2000
|
|
