3SWJ
 
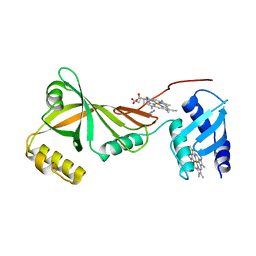 | | Crystal structure of Campylobacter jejuni ChuZ | | Descriptor: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | Authors: | Hu, Y. | | Deposit date: | 2011-07-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
6AC0
 
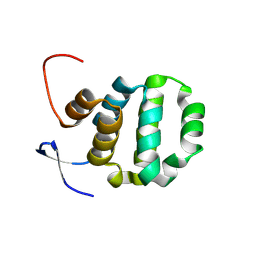 | | Crystal structure of TRADD death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6AC5
 
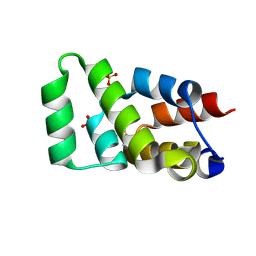 | | Crystal structure of RIPK1 death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6ACI
 
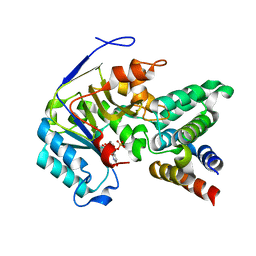 | | Crystal structure of EPEC effector NleB in complex with FADD death domain | | Descriptor: | FAS-associated death domain protein, MANGANESE (II) ION, T3SS secreted effector NleB homolog, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-26 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
5Z2C
 
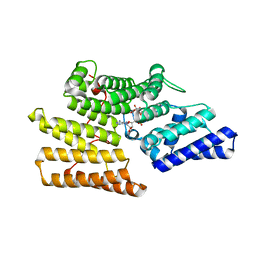 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
3AFK
 
 | |
2Q8Y
 
 | |
5O2F
 
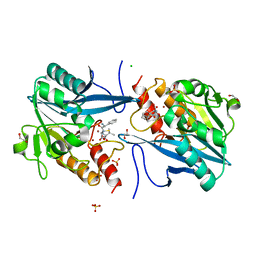 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5O2E
 
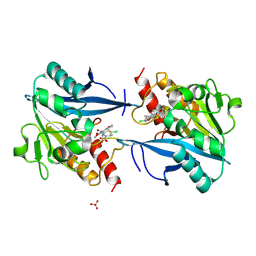 | | Crystal structure of NDM-1 in complex with hydrolyzed cefuroxime - new refinement | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
