3JUA
 
 | |
1YHN
 
 | | Structure basis of RILP recruitment by Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | Authors: | Wu, M, Wang, T, Hong, W, Song, H. | | Deposit date: | 2005-01-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
207L
 
 | | MUTANT HUMAN LYSOZYME C77A | | Descriptor: | LYSOZYME, N-ACETYL-L-CYSTEINE | | Authors: | Matsushima, M, Song, H. | | Deposit date: | 1996-03-26 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A role of PDI in the reductive cleavage of mixed disulfides.
J.Biochem.(Tokyo), 120, 1996
|
|
2A6T
 
 | |
1XML
 
 | | Structure of human Dcps | | Descriptor: | PHOSPHATE ION, heat shock-like protein 1 | | Authors: | Chen, N, Song, H. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human DcpS in ligand-free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping.
J.Mol.Biol., 347, 2005
|
|
208L
 
 | | MUTANT HUMAN LYSOZYME C77A | | Descriptor: | CYSTEINE, LYSOZYME | | Authors: | Matsushima, M, Song, H. | | Deposit date: | 1996-03-26 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A role of PDI in the reductive cleavage of mixed disulfides.
J.Biochem.(Tokyo), 120, 1996
|
|
4B6H
 
 | |
8GV5
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin A/swine/Guangdong/104/2013 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV4
 
 | | A human broadly neutralizing influenza A hemagglutinin stem-specific antibody PN-SIA28 | | Descriptor: | PN-SIA28 heavy chain, PN-SIA28 light chain | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV6
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin H14 (A/long-tailed duck/Wisconsin/10OS3912/2010) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin H14-HA1, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV7
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin H18 A/flat-faced bat/Peru/033/2010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin H18-HA1, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
4B3F
 
 | | crystal structure of Ighmbp2 helicase | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, PHOSPHATE ION | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
3E20
 
 | | Crystal structure of S.pombe eRF1/eRF3 complex | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
3EIQ
 
 | | Crystal structure of Pdcd4-eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Programmed cell death protein 4 | | Authors: | Loh, P.G, Cheng, Z, Song, H. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
4LL9
 
 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
4LLA
 
 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
3E1Y
 
 | | Crystal structure of human eRF1/eRF3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
3D45
 
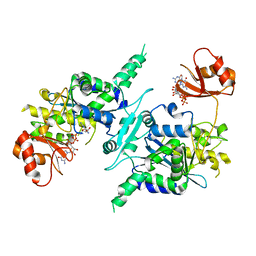 | | Crystal structure of mouse PARN in complex with m7GpppG | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | Authors: | Wu, M, Song, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease.
Structure, 17, 2009
|
|
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZN
 
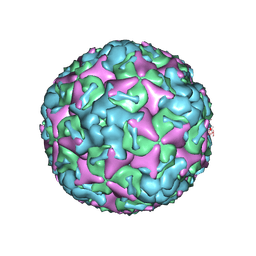 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4B3G
 
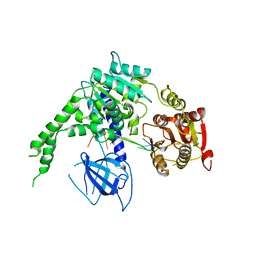 | | crystal structure of Ighmbp2 helicase in complex with RNA | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, RNA (5'-(AP*AP*AP*AP*AP*AP*AP*AP*AP)-3') | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
7BZO
 
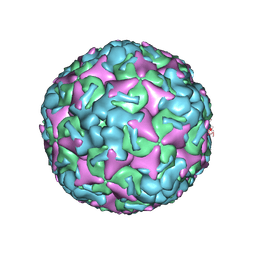 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZU
 
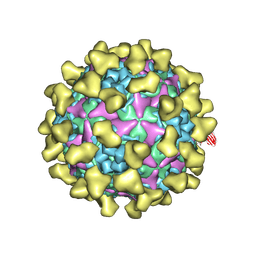 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
1SV0
 
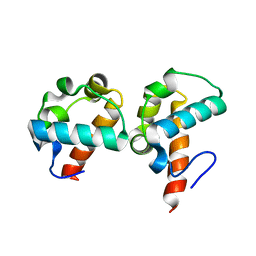 | | Crystal Structure Of Yan-SAM/Mae-SAM Complex | | Descriptor: | Ets DNA-binding protein pokkuri, modulator of the activity of Ets CG15085-PA | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
