3MCI
 
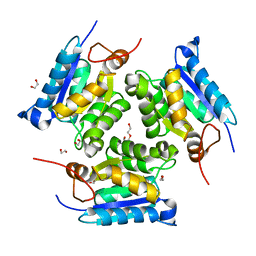 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3MCJ
 
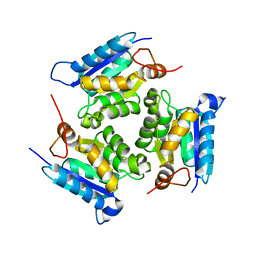 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) other form from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1TUX
 
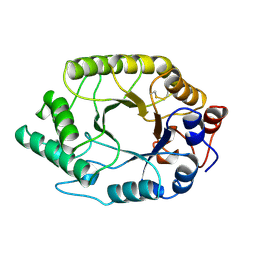 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS | | Descriptor: | XYLANASE | | Authors: | Natesh, R, Bhanumoorthy, P, Vithayathil, P.J, Sekar, K, Ramakumar, S, Viswamitra, M.A. | | Deposit date: | 1998-10-29 | | Release date: | 1999-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and proposed amino acid sequence of a thermostable xylanase from Thermoascus aurantiacus.
J.Mol.Biol., 288, 1999
|
|
1TH3
 
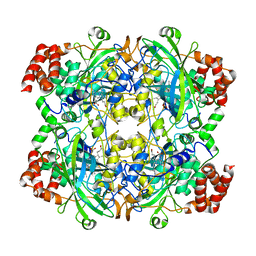 | | Crystal structure of NADPH depleted bovine live catalase complexed with cyanide | | Descriptor: | CYANIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1TH4
 
 | | crystal structure of NADPH depleted bovine liver catalase complexed with 3-amino-1,2,4-triazole | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1TGU
 
 | | The crystal structure of bovine liver catalase without NADPH | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-05-31 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1VEQ
 
 | | Mycobacterium smegmatis Dps Hexagonal form | | Descriptor: | FE (III) ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-03 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
1TH2
 
 | | crystal structure of NADPH depleted bovine liver catalase complexed with azide | | Descriptor: | AZIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1VEL
 
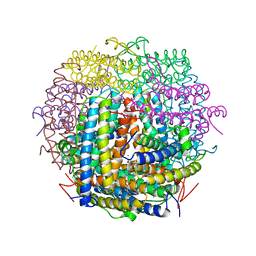 | | Mycobacterium smegmatis Dps tetragonal form | | Descriptor: | CADMIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-01 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
1VEI
 
 | | Mycobacterium smegmatis Dps | | Descriptor: | FE (III) ION, SULFATE ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray Analysis of Mycobacterium smegmatis Dps and a Comparative Study Involving Other Dps and Dps-like Molecules
J.Mol.Biol., 339, 2004
|
|
3U54
 
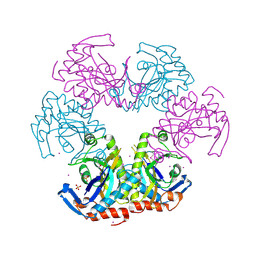 | | Crystal structure (Type-1) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | Descriptor: | 1,4-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2011-10-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
1MKS
 
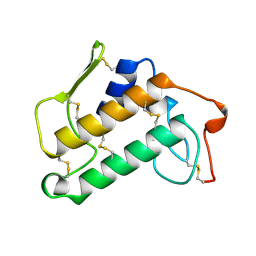 | |
1FDK
 
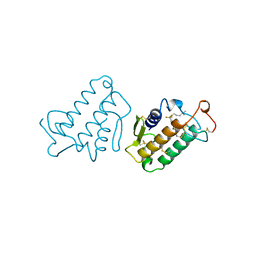 | | CARBOXYLIC ESTER HYDROLASE (PLA2-MJ33 INHIBITOR COMPLEX) | | Descriptor: | 1-DECYL-3-TRIFLUORO ETHYL-SN-GLYCERO-2-PHOSPHOMETHANOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-09-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the complex of bovine pancreatic phospholipase A2 with the inhibitor 1-hexadecyl-3-(trifluoroethyl)-sn-glycero-2-phosphomethanol,.
Biochemistry, 36, 1997
|
|
1KVW
 
 | |
1KVX
 
 | |
1KVY
 
 | |
1MKV
 
 | |
1UNE
 
 | |
1MKT
 
 | |
1MKU
 
 | |
4R6R
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6Q
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
