6C14
 
 | |
5IVX
 
 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Jiang, J, Margulies, D. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
5J60
 
 | | Structure of a thioredoxin reductase from Gloeobacter violaceus | | Descriptor: | CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, TETRAETHYLENE GLYCOL, ... | | Authors: | Buey, R.M, de Pereda, J.M, Balsera, M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Member of the Thioredoxin Reductase Family from Early Oxygenic Photosynthetic Organisms.
Mol Plant, 10, 2017
|
|
6FLP
 
 | |
3SAQ
 
 | |
3SAM
 
 | |
3U64
 
 | | The Crystal Structure of Tat-T (Tp0956) | | Descriptor: | Protein TP_0956, SULFATE ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3U65
 
 | | The Crystal Structure of Tat-P(T) (Tp0957) | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, Tp33 protein | | Authors: | Brautigam, C.A, Tomchick, D.R, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3H6H
 
 | |
3G3H
 
 | |
3G3G
 
 | |
3G3K
 
 | |
3G3J
 
 | |
3G3I
 
 | |
3G3F
 
 | |
3H6G
 
 | |
6GND
 
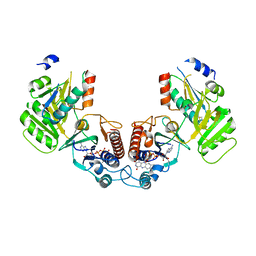 | | Crystal structure of the complex of a Ferredoxin-Flavin Thioredoxin Reductase and a Thioredoxin from Clostridium acetobutylicum at 2.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GNB
 
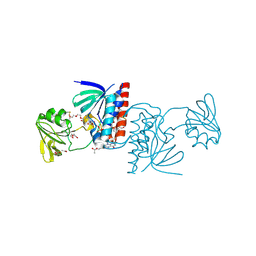 | | Crystal structure of a Ferredoxin-Flavin Thioredoxin Reductase from Clostridium acetobutylicum at 1.9 A resolution | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GN9
 
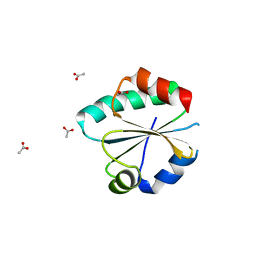 | |
6GNC
 
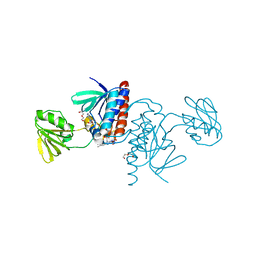 | |
3QLV
 
 | |
6GNA
 
 | | Crystal structure of a Ferredoxin-Flavin Thioredoxin Reductase from Clostridium acetobutylicum at 1.3 A resolution | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.295 Å) | | Cite: | Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3QLT
 
 | |
6FLQ
 
 | |
