3AEG
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with N-Biphenyl-3-yl-2-iodo-benzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with N-Biphenyl-3-yl-2-iodo-benzamide
To be Published
|
|
3W3E
 
 | |
3AE8
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with N-(3-Isopropoxy-phenyl)-2-trifluoromethylbenzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with N-(3-Isopropoxy-phenyl)-2-trifluoromethylbenzamide
To be Published
|
|
3AE5
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Methyl-N-(3-isopropoxy-phenyl)-benzamide | | Descriptor: | 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Methyl-N-(3-isopropoxy-phenyl)-benzamide
To be Published
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
2D1Q
 
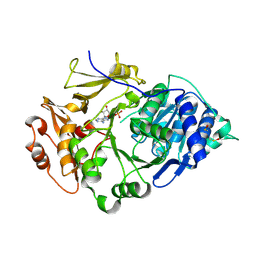 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with MgATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1R
 
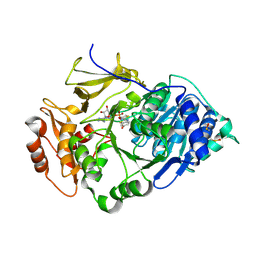 | | Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP | | Descriptor: | 2-(6-HYDROXY-1,3-BENZOTHIAZOL-2-YL)-1,3-THIAZOL-4(5H)-ONE, ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1S
 
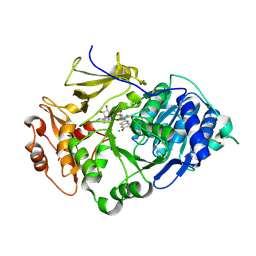 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1T
 
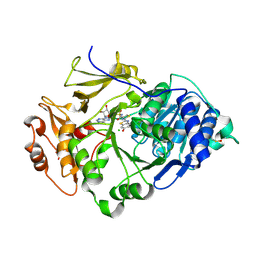 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2EJL
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
2EJ6
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound D-proline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
2EII
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-valine and NAD. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD.
To be Published
|
|
2EHQ
 
 | | Crystal analysis of 1-pyrroline-5-carboxylate dehydrogenase from thermus with bound NADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New insights into the binding mode of coenzymes: structure of Thermus thermophilus Delta1-pyrroline-5-carboxylate dehydrogenase complexed with NADP+.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2EIW
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-proline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
2EIT
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-alanine and NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
2EJD
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-alanine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
4ZHX
 
 | | Novel binding site for allosteric activation of AMPK | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-2, ... | | Authors: | Langendorf, C.G, Ngoei, K.R, Issa, S.M.A, Ling, N, Gorman, M.A, Parker, M.W, Sakamoto, K, Scott, J.W, Oakhill, J.S, Kemp, B.E. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Nat Commun, 7, 2016
|
|
2H9U
 
 | | Crystal structure of the archaea specific DNA binding protein | | Descriptor: | 1,2-ETHANEDIOL, DNA/RNA-binding protein Alba 2 | | Authors: | Kumarevel, T.S, Sakamoto, K, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the archaea specific DNA binding protein from Aeropyrum pernix K1
To be Published
|
|
6KK8
 
 | | XN joint refinement of manganese catalase from Thermus Thermophilus HB27 | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (III) ION, OXYGEN ATOM, ... | | Authors: | Yamada, T, Yano, N, Kusaka, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.37 Å), X-RAY DIFFRACTION | | Cite: | Single-crystal time-of-flight neutron Laue methods: application to manganese catalase from Thermus thermophilus HB27
J.Appl.Crystallogr., 2019
|
|
2J53
 
 | | Solution Structure of GB1 domain Protein G and low and high pressure. | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wilton, D.J, Tunnicliffe, R.B, Kamatari, Y.O, Akasaka, K, Williamson, M.P. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pressure-Induced Changes in the Solution Structure of the Gb1 Domain of Protein G.
Proteins, 71, 2008
|
|
7VEI
 
 | | Neutron structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1V81
 
 | |
5YVQ
 
 | | Complex of Mu phage tail fiber and its chaperone | | Descriptor: | GLYCEROL, Tail fiber assembly protein U, Tail fiber protein S | | Authors: | Takeda, S, Sakai, K, Iwazaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-11-27 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres.
Nat Microbiol, 4, 2019
|
|
1V80
 
 | |
1GXV
 
 | |
