3FU7
 
 | | Melanocarpus albomyces laccase crystal soaked (4 sec) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxycyclohexa-2,5-diene-1,4-dione, 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
3PPS
 
 | | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria--common structural features of asco-laccases.
Febs J., 278, 2011
|
|
3FU9
 
 | | Melanocarpus albomyces laccase crystal soaked (20 min) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxybenzene-1,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
3FU8
 
 | | Melanocarpus albomyces laccase crystal soaked (10 sec) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
3QPK
 
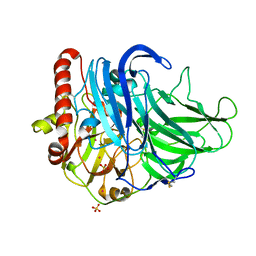 | | Probing oxygen channels in Melanocarpus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2011-02-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Dioxygen Route in Melanocarpus albomyces Laccase with Pressurized Xenon Gas.
Biochemistry, 50, 2011
|
|
3ZZQ
 
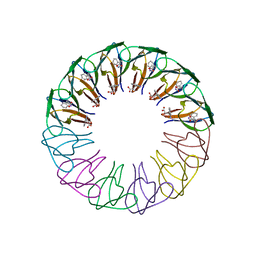 | | Engineered 12-subunit Bacillus subtilis trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
2R56
 
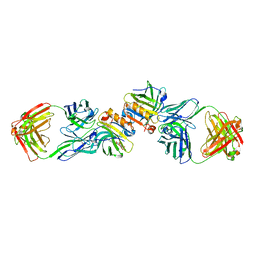 | | Crystal Structure of a Recombinant IgE Fab Fragment in Complex with Bovine Beta-Lactoglobulin Allergen | | Descriptor: | Beta-lactoglobulin, DODECYL-BETA-D-MALTOSIDE, IgE Fab Fragment, ... | | Authors: | Niemi, M, Kallio, J.M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen.
Structure, 15, 2007
|
|
3ZZS
 
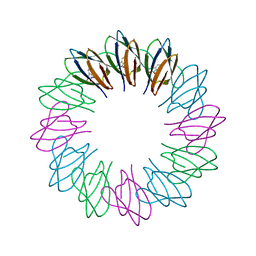 | | Engineered 12-subunit Bacillus stearothermophilus trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZZL
 
 | | Bacillus halodurans trp RNA-binding attenuation protein (TRAP): a 12- subunit assembly | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
6FHC
 
 | |
6GS3
 
 | |
6FG4
 
 | |
6FGR
 
 | |
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5A2T
 
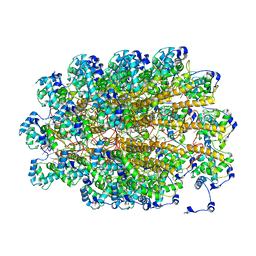 | | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses | | Descriptor: | BAMBOO MOSAIC VIRUS, COAT PROTEIN | | Authors: | DiMaio, F, Chen, C.C, Yu, X, Frenz, B, Hsu, Y.H, Lin, N.S, Egelman, E.H. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5J72
 
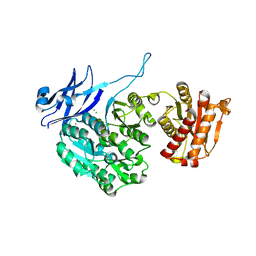 | | Cwp6 from Clostridium difficile | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
5J6Q
 
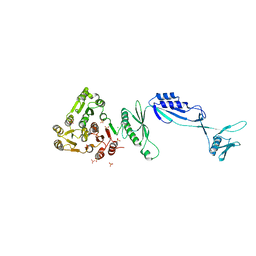 | | Cwp8 from Clostridium difficile | | Descriptor: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
5BWW
 
 | |
5BWU
 
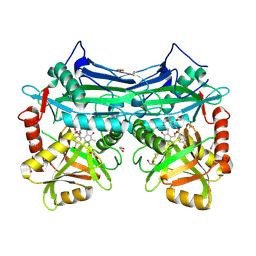 | | X-RAY CRYSTAL STRUCTURE AT 2.17A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TRIAZOLOPYRIMIDINONE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4-bromobenzyl)amino]-5-propyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2015-06-08 | | Release date: | 2015-07-01 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Discovery of in Vivo Active Mitochondrial Branched-Chain Aminotransferase (BCATm) Inhibitors by Hybridizing Fragment and HTS Hits.
J.Med.Chem., 58, 2015
|
|
6W0V
 
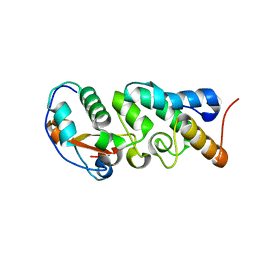 | | The Crystal Structure of the Mutant Nuclease Domain of Pyocin S8 with its Cognate Immunity Protein | | Descriptor: | Bacteriocin immunity protein, Pyocin S8 | | Authors: | Turano, H, Gomes, F, Domingos, R.M, Netto, L.E.S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Molecular Structure and Functional Analysis of Pyocin S8 from Pseudomonas aeruginosa Reveals the Essential Requirement of a Glutamate Residue in the H-N-H Motif for DNase Activity.
J.Bacteriol., 202, 2020
|
|
7BYF
 
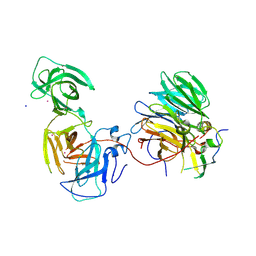 | | The crystal structure of mouse ORF10-Rae1-Nup98 complex | | Descriptor: | 10 protein, MERCURY (II) ION, Peptidase S59 domain-containing protein, ... | | Authors: | Gao, P, Feng, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6G8D
 
 | |
6G9G
 
 | |
6G8E
 
 | |
6G8C
 
 | |
