1RKM
 
 | | STRUCTURE OF OPPA | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
9G17
 
 | | Structure of PslG with a covalently- bound pentasaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, PslG, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2024-07-09 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bespoke Activity-Based Probes Reveal that the Pseudomonas aeruginosa Endoglycosidase, PslG, Is an Endo-beta-glucanase.
J.Am.Chem.Soc., 147, 2025
|
|
9G18
 
 | | Structure of PslG with an iminosugar inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, PslG, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2024-07-09 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bespoke Activity-Based Probes Reveal that the Pseudomonas aeruginosa Endoglycosidase, PslG, Is an Endo-beta-glucanase.
J.Am.Chem.Soc., 147, 2025
|
|
2AAA
 
 | | CALCIUM BINDING IN ALPHA-AMYLASES: AN X-RAY DIFFRACTION STUDY AT 2.1 ANGSTROMS RESOLUTION OF TWO ENZYMES FROM ASPERGILLUS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1991-02-27 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus.
Biochemistry, 29, 1990
|
|
1LFH
 
 | | MOLECULAR REPLACEMENT SOLUTION OF THE STRUCTURE OF APOLACTOFERRIN, A PROTEIN DISPLAYING LARGE-SCALE CONFORMATIONAL CHANGE | | Descriptor: | CHLORIDE ION, LACTOFERRIN | | Authors: | Anderson, B.F, Baker, E.N, Norris, G.E. | | Deposit date: | 1991-09-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular replacement solution of the structure of apolactoferrin, a protein displaying large-scale conformational change.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1BNI
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
5CNA
 
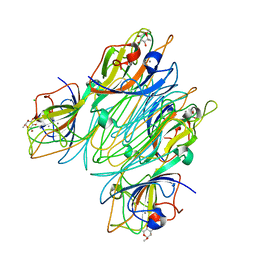 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1EVR
 
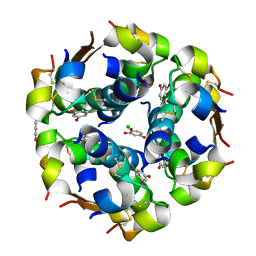 | | The structure of the resorcinol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, RESORCINOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-20 | | Release date: | 2000-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EV6
 
 | | Structure of the monoclinic form of the M-cresol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-19 | | Release date: | 2000-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | R6 Hexameric Insulin Complexed with m-Cresol or Resorcinol
Biochem.Biophys.Res.Commun., 56, 2000
|
|
1EV3
 
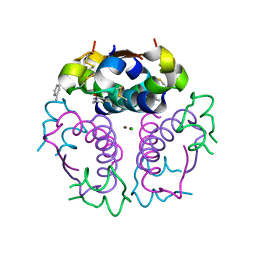 | | Structure of the rhombohedral form of the M-cresol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-19 | | Release date: | 2000-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1IZB
 
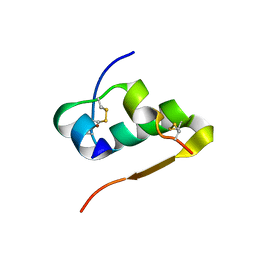 | |
3W80
 
 | |
3W7Y
 
 | |
1IZA
 
 | |
1MNI
 
 | |
1TYL
 
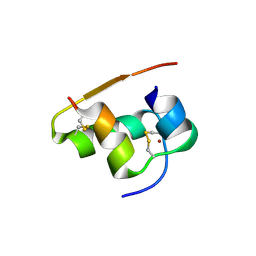 | |
1TYM
 
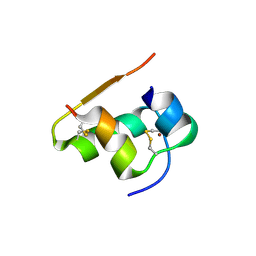 | |
1UTD
 
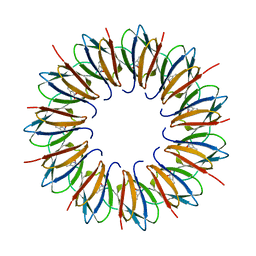 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a 63-nucleotide RNA molecule containing GAGUUU repeats | | Descriptor: | 5'-R(*GP*UP*UP*UP*GP*AP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
1LPH
 
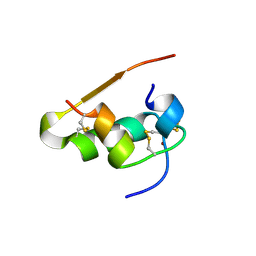 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
1MNH
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Davies, G.J, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1G7A
 
 | | 1.2 A structure of T3R3 human insulin at 100 K | | Descriptor: | ACETONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Smith, G.D, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-11-09 | | Release date: | 2001-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Phase changes in T(3)R(3)(f) human insulin: temperature or pressure induced?
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1MNJ
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNK
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
2A3G
 
 | |
1QKA
 
 | |
