1YDD
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDA
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
2Z6M
 
 | |
2ZAV
 
 | |
4MYL
 
 | |
4MYF
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase(oxidized) with Mn2+2 at pH 6.0 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4MYK
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase (oxidized) with Mn2+2 at pH 8.5 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4LZ3
 
 | | F95H Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, HYDROGENPHOSPHATE ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZ0
 
 | | A236G Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
3BUV
 
 | | Crystal structure of human Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and HEPES. Resolution: 1.35 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-03 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
4LTZ
 
 | | F95M Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LUU
 
 | | V329A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LXW
 
 | | L72V Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
3BNX
 
 | |
4LTV
 
 | | Crystal structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2) | | Descriptor: | Epi-isozizaene synthase, SULFATE ION | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZC
 
 | | W325F Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
3BV7
 
 | | Crystal structure of Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and glycerol. Resolution: 1.79 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
4MXR
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase with Mn2+2 | | Descriptor: | Formiminoglutamase, GLYCEROL, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.-J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4MYN
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase N114H variant with Mn2+2 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
3BUR
 
 | |
3CYU
 
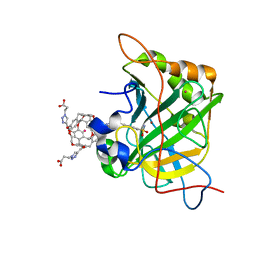 | | Human Carbonic Anhydrase II complexed with Cryptophane biosensor and xenon | | Descriptor: | Carbonic anhydrase 2, MoMo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-y l)propanoic acid]-cryptophane-A, PoPo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-yl)propanoic acid]-cryptophane-A, ... | | Authors: | Aaron, J.A, Jude, K.M, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2008-04-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II.
J.Am.Chem.Soc., 130, 2008
|
|
3BNY
 
 | | Crystal structure of aristolochene synthase complexed with 2-fluorofarnesyl diphosphate (2F-FPP) | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Christianson, D.W. | | Deposit date: | 2007-12-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis
J.Biol.Chem., 283, 2008
|
|
3COT
 
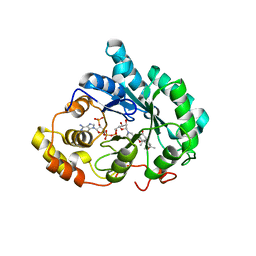 | | Crystal structure of human liver delta(4)-3-ketosteroid 5beta-reductase (akr1d1) in complex with progesterone and nadp. Resolution: 2.03 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
7KUV
 
 | |
