3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3FG7
 
 | |
3F6F
 
 | |
3FFN
 
 | |
3F5J
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F63
 
 | |
3F6D
 
 | | Crystal Structure of a Genetically Modified Delta Class GST (adGSTD4-4) from Anopheles dirus, F123A, in Complex with S-Hexyl Glutathione | | Descriptor: | Glutathione transferase GST1-4, S-HEXYLGLUTATHIONE | | Authors: | Wongsantichon, J, Robinson, R.C, Ketterman, A.J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural contributions of delta class glutathione transferase active-site residues to catalysis
Biochem.J., 428, 2010
|
|
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F4V
 
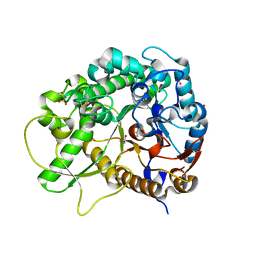 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3FFK
 
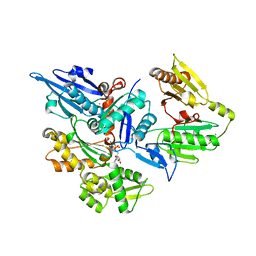 | | Crystal structure of human Gelsolin domains G1-G3 bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, actin, ... | | Authors: | Chumnarnsilpa, S, Robinson, R.C, Burtnick, L.D. | | Deposit date: | 2008-12-03 | | Release date: | 2009-10-06 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G7I
 
 | |
3G7J
 
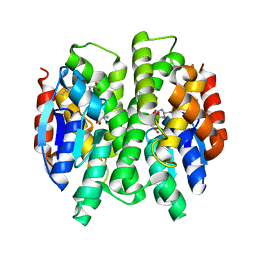 | | Crystal Structure of a Genetically Modified Delta Class GST (adGSTD4-4) from Anopheles dirus, Y119E, in Complex with S-Hexyl Glutathione | | Descriptor: | Glutathione transferase GST1-4, S-HEXYLGLUTATHIONE | | Authors: | Wongsantichon, J, Robinson, R.C, Ketterman, A.J. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural contributions of delta class glutathione transferase active-site residues to catalysis
Biochem.J., 428, 2010
|
|
3GH6
 
 | |
3HBT
 
 | | The structure of native G-actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Wang, H, Robinson, R.C, Burtnick, L.D. | | Deposit date: | 2009-05-05 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of native G-actin
Cytoskeleton (Hoboken), 67, 2010
|
|
3LK4
 
 | | Crystal structure of CapZ bound to the uncapping motif from CD2AP | | Descriptor: | CD2-associated protein, F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2 | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LK2
 
 | | Crystal structure of CapZ bound to the uncapping motif from CARMIL | | Descriptor: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Aguda, A.H, Larsson, M, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LK3
 
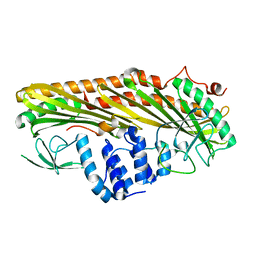 | | Crystal structure of CapZ bound to the CPI and CSI uncapping motifs from CARMIL | | Descriptor: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MAK
 
 | |
7CKB
 
 | | Simplified Alpha-Carboxysome, T=3 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7CKC
 
 | | Simplified Alpha-Carboxysome, T=4 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7DHQ
 
 | | Structure of Halothiobacillus neapolitanus Microcompartments Protein CsoS1D | | Descriptor: | Microcompartments protein | | Authors: | Xue, B, Tan, Y.Q, Ali, S, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7EBI
 
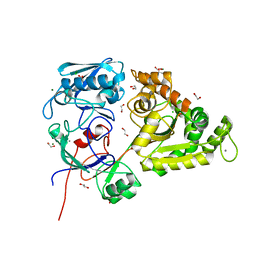 | | Chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitotetraose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
7EBM
 
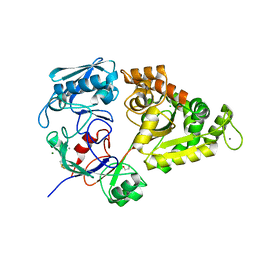 | | W363A mutant of Chitin-specific solute binding protein from Vibrio harveyi in complex with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
1D0N
 
 | | THE CRYSTAL STRUCTURE OF CALCIUM-FREE EQUINE PLASMA GELSOLIN. | | Descriptor: | HORSE PLASMA GELSOLIN | | Authors: | Burtnick, L.D, Robinson, R, Li, C. | | Deposit date: | 1999-09-13 | | Release date: | 1999-09-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation.
Cell(Cambridge,Mass.), 90, 1997
|
|
8JBP
 
 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.45 angstroms resolution with an arsenic atom bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Leartsakulpanich, U, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Robinson, R.C, Zhou, Y, Mungthin, M, Leelayoova, S, Saehlee, S, Choowongkomon, K. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
