2Y6P
 
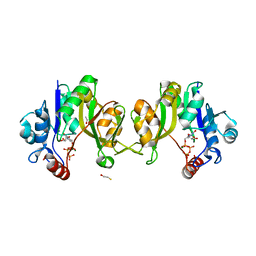 | | Evidence for a Two-Metal-Ion-Mechanism in the Kdo- Cytidylyltransferase KdsB | | Descriptor: | 3-DEOXY-MANNO-OCTULOSONATE CYTIDYLYLTRANSFERASE, BETA-MERCAPTOETHANOL, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Schmidt, H, Mesters, J.R, Mamat, U, Hilgenfeld, R. | | Deposit date: | 2011-01-25 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence for a Two-Metal-Ion Mechanism in the Cytidyltransferase Kdsb, an Enzyme Involved in Lipopolysaccharide Biosynthesis.
Plos One, 6, 2011
|
|
5NH0
 
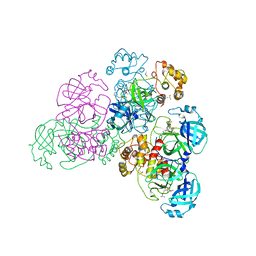 | |
2WCT
 
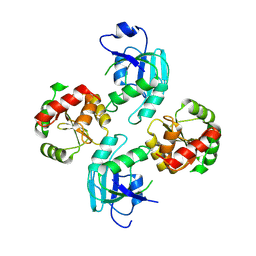 | | human SARS coronavirus unique domain (triclinic form) | | Descriptor: | NON-STRUCTURAL PROTEIN 3 | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
3UB0
 
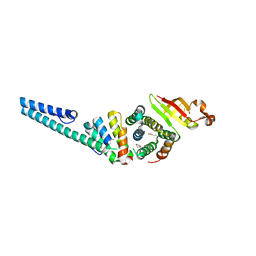 | | Crystal structure of the nonstructural protein 7 and 8 complex of Feline Coronavirus | | Descriptor: | Non-structural protein 6, nsp6,, Non-structural protein 7, ... | | Authors: | Xiao, Y, Hilgenfeld, R, Ma, Q. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity.
J.Virol., 86, 2012
|
|
1P9S
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replicase polyprotein 1ab | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
1P9U
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHQ-VNSTLQ-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION, ... | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
3EJF
 
 | |
1DPW
 
 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME IN COMPLEX WITH MPD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | Deposit date: | 1999-12-28 | | Release date: | 2000-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DPX
 
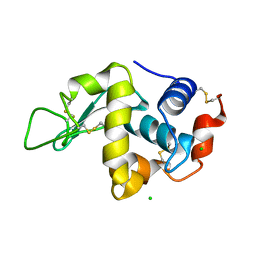 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | Deposit date: | 1999-12-28 | | Release date: | 2000-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3PV2
 
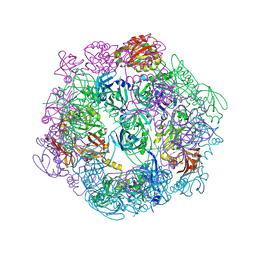 | |
3PV4
 
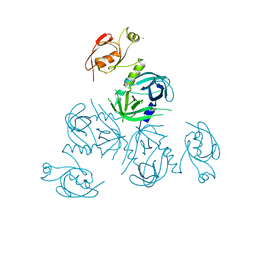 | | Structure of Legionella fallonii DegQ (Delta-PDZ2 variant) | | Descriptor: | CADMIUM ION, DegQ | | Authors: | Wrase, R, Scott, H, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2010-12-06 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PV5
 
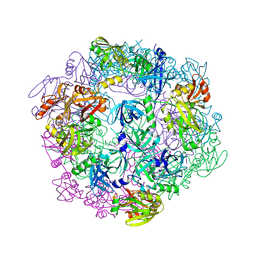 | |
3PV3
 
 | | Structure of Legionella fallonii DegQ (S193A variant) | | Descriptor: | DegQ, Substrate peptide (Poly-Ala) | | Authors: | Wrase, R, Scott, H, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2010-12-06 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P31
 
 | |
3O5Z
 
 | | Crystal structure of the SH3 domain from p85beta subunit of phosphoinositide 3-kinase (PI3K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Chen, S, Xiao, Y, Ponnusamy, R, Tan, J, Lei, J, Hilgenfeld, R. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-ray structure of the SH3 domain of the phosphoinositide 3-kinase p85 beta subunit
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1FD9
 
 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | Authors: | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
2C0Y
 
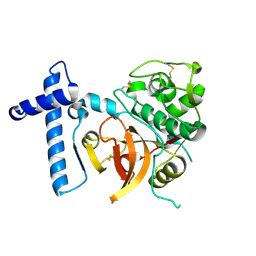 | | THE CRYSTAL STRUCTURE OF A CYS25ALA MUTANT OF HUMAN PROCATHEPSIN S | | Descriptor: | PROCATHEPSIN S | | Authors: | Kaulmann, G, Palm, G.J, Schilling, K, Hilgenfeld, R, Wiederanders, B. | | Deposit date: | 2005-09-08 | | Release date: | 2006-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Cys25 -> Ala Mutant of Human Procathepsin S Elucidates Enzyme-Prosequence Interactions.
Protein Sci., 15, 2006
|
|
1DG1
 
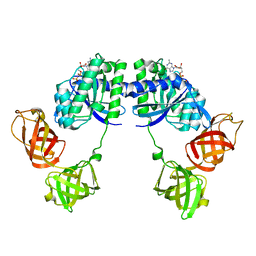 | | WHOLE, UNMODIFIED, EF-TU(ELONGATION FACTOR TU). | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Abel, K, Yoder, M, Hilgenfeld, R, Jurnak, F. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alpha to beta conformational switch in EF-Tu.
Structure, 4, 1996
|
|
3TNT
 
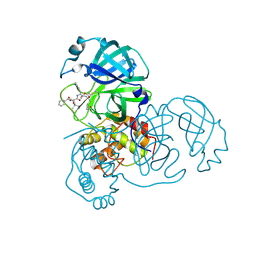 | |
3SNA
 
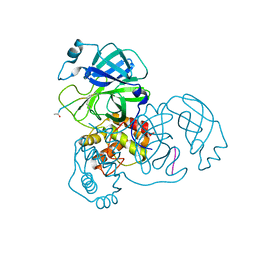 | |
3SZN
 
 | |
3TIU
 
 | | Crystal structure of SARS coronavirus main protease complexed with an alpha,beta-unsaturated ethyl ester inhibitor SG82 | | Descriptor: | DIMETHYL SULFOXIDE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE, SARS coronavirus main protease | | Authors: | Zhu, L, Hilgenfeld, R. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of SARS-Cov main protease complexed with a series of peptidic unsaturated esters
To be Published
|
|
3TIT
 
 | |
3TNS
 
 | | Crystal structure of SARS coronavirus main protease complexed with an alpha, beta-unsaturated ethyl ester inhibitor SG83 | | Descriptor: | ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE, SARS coronavirus main protease | | Authors: | Zhu, L, Hilgenfeld, R. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of SARS-Cov main protease complexed with a series of peptidic unsaturated esters
To be Published
|
|
1UJ1
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
