9F9K
 
 | | Crystal structure of MUS81-EME1 bound by compound 15. | | Descriptor: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F99
 
 | | Crystal structure of MUS81-EME1 bound by compound 10. | | Descriptor: | 2-(4-chlorophenyl)-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F9A
 
 | | Crystal structure of MUS81-EME1 bound by compound 12. | | Descriptor: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F9L
 
 | | Crystal structure of MUS81-EME1 bound by compound 16. | | Descriptor: | 2-[2-[4-(cyanomethyl)phenyl]phenyl]-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
4OBX
 
 | | Crystal structure of yeast Coq5 in the apo form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OBW
 
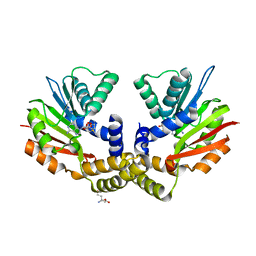 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7YTJ
 
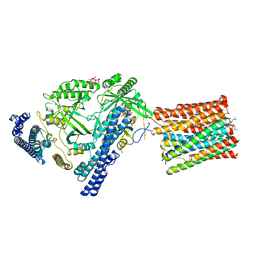 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
4X36
 
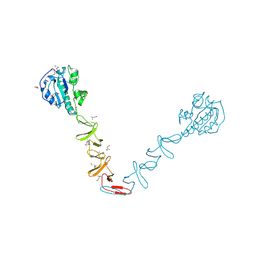 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XSP
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP | | Descriptor: | Alr3699 protein, GLYCEROL, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSR
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP-glucose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alr3699 protein, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSO
 
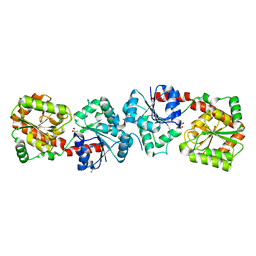 | | Crystal structure of apo-form Alr3699/HepE from Anabaena sp. strain PCC 7120 | | Descriptor: | Alr3699 protein, GLYCEROL | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSU
 
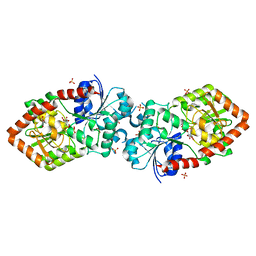 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP and glucose | | Descriptor: | Alr3699 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
8H08
 
 | |
8H07
 
 | |
4U8Z
 
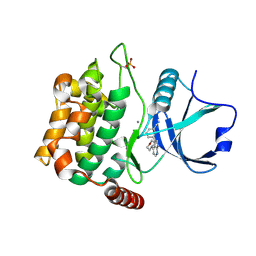 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06447475) | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, MANGANESE (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
2WIB
 
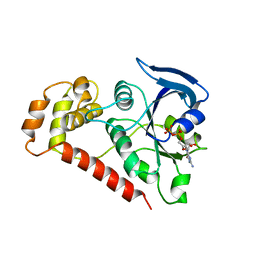 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GDP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2WIA
 
 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in Apo Form | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2WIC
 
 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GMPPNP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
6K9E
 
 | |
7YMK
 
 | | Estrogen Receptor Alpha Ligand Binding Domain C381S C417S Y537S Mutant in Complex with an Covalent Selective Estrogen Receptor Degrader 29c and GRIP Peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Estrogen receptor, Grip peptide, ... | | Authors: | Min, J, Hu, H.B, Yang, Y, Dong, C.E, Zhou, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-07-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of novel covalent selective estrogen receptor degraders against endocrine-resistant breast cancer.
Acta Pharm Sin B, 13, 2023
|
|
8QJR
 
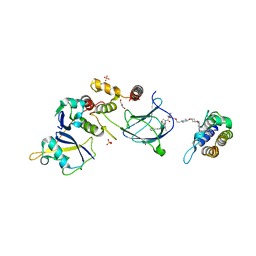 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJS
 
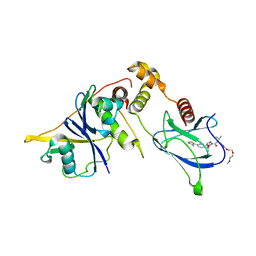 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJT
 
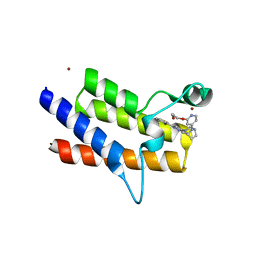 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
5OV7
 
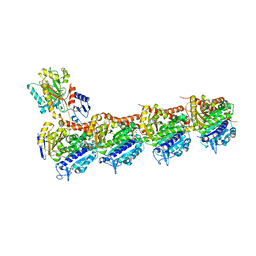 | | tubulin - rigosertib complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M, Jost, M. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent.
Mol. Cell, 68, 2017
|
|
8FR6
 
 | | Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
