8SHA
 
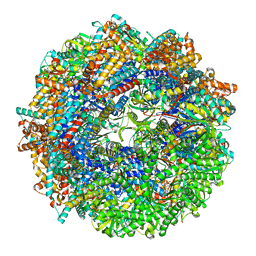 | | CCT-G beta 5 complex closed state 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHO
 
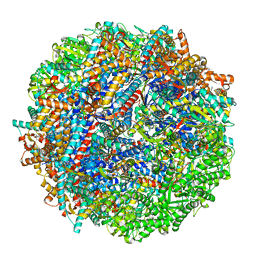 | | CCT G beta 5 complex close state 11 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHE
 
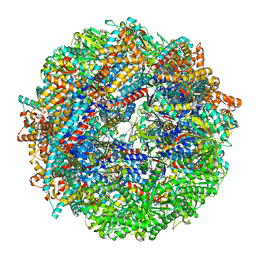 | | CCT-G beta 5 complex closed state 8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHL
 
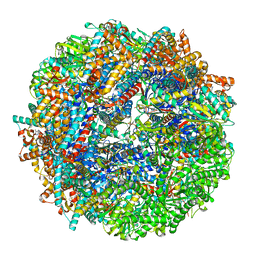 | | CCT G beta 5 complex closed state 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SH9
 
 | | CCT G beta 5 complex best PhLP1 class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHF
 
 | | CCT-G beta 5 complex closed state 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHT
 
 | | CCT G beta 5 complex closed state 14 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHN
 
 | | CCT-G beta 5 complex closed state 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHQ
 
 | | CCT G beta 5 complex closed state 12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHG
 
 | | CCT G beta 5 complex closed state 9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8TOX
 
 | |
2L0M
 
 | | DsbB2 peptide structure in 100% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0O
 
 | | DsbB3 peptide structure in 100% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
5DLI
 
 | | Corkscrew assembly of SOD1 residues 28-38 | | Descriptor: | GLYCEROL, IODIDE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2015-09-05 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2L0L
 
 | | DsbB2 peptide structure in 70% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0N
 
 | | DsbB3 peptide structure in 70% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
3EBG
 
 | | Structure of the M1 Alanylaminopeptidase from malaria | | Descriptor: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2008-08-27 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EBH
 
 | | Structure of the M1 Alanylaminopeptidase from malaria complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2008-08-27 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5WMJ
 
 | |
5WOR
 
 | |
3EBI
 
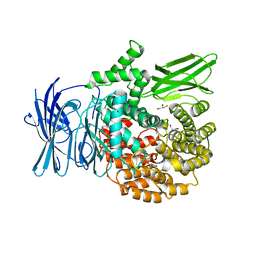 | | Structure of the M1 Alanylaminopeptidase from malaria complexed with the phosphinate dipeptide analog | | Descriptor: | (2S)-3-[(R)-[(1S)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2008-08-27 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4F88
 
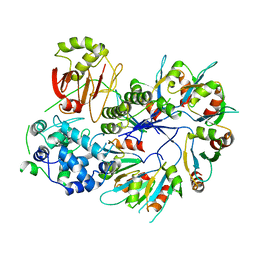 | | X-ray Crystal Structure of PlyC | | Descriptor: | PlyCA, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F87
 
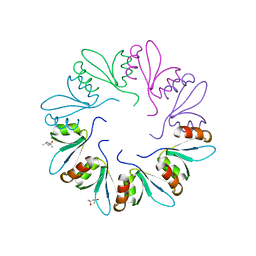 | | X-ray Crystal Structure of PlyCB | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MBB
 
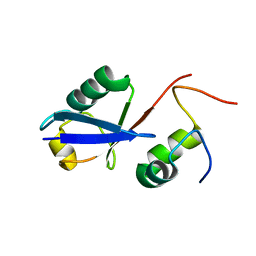 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
