7LR5
 
 | |
7LSJ
 
 | |
7LRR
 
 | |
7LSL
 
 | |
7LRA
 
 | |
7LRV
 
 | |
7LSN
 
 | |
7LRB
 
 | |
7LV4
 
 | |
7LV1
 
 | | Cu-bound crystal structure of the engineered cyt cb562 variant, DiCyt2, crystallized in the presence of Cu(II) | | Descriptor: | CALCIUM ION, COPPER (II) ION, HEME C, ... | | Authors: | Choi, T.S, Tezcan, F.A. | | Deposit date: | 2021-02-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Overcoming universal restrictions on metal selectivity by protein design.
Nature, 603, 2022
|
|
6O7M
 
 | | Nitrogenase MoFeP mutant F99Y from Azotobacter vinelandii in the indigo carmine oxidized state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
6O7O
 
 | |
6O7S
 
 | |
6O7L
 
 | |
6O7R
 
 | | Nitrogenase MoFeP mutant F99Y, S188A from Azotobacter vinelandii in the dithionite reduced state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
6O7P
 
 | | Nitrogenase MoFeP mutant F99Y from Azotobacter vinelandii in the dithionite reduced state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
6O7Q
 
 | |
6OT9
 
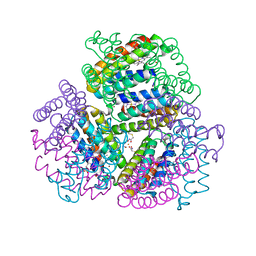 | | Bimetallic dodecameric cage design 1 (BMC1) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT4
 
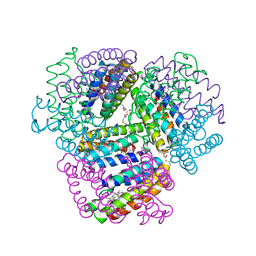 | | Bimetallic dodecameric cage design 2 (BMC2) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT7
 
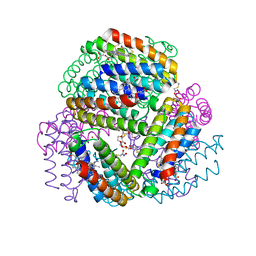 | | Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT8
 
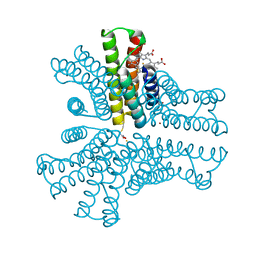 | | Bimetallic hexameric cage design 4 (BMC4) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OVH
 
 | | Cryo-EM structure of Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Subramanian, R.H, Yan, X, Alberstein, R.G, Tezcan, F.A. | | Deposit date: | 2019-05-07 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6O7N
 
 | |
3DE8
 
 | | Crystal Structure of a Dimeric Cytochrome cb562 Assembly Induced by Copper Coordination | | Descriptor: | CALCIUM ION, COPPER (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Salgado, E.N, Lewis, R.A, Rheingold, A.L, Tezcan, F.A. | | Deposit date: | 2008-06-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through Cu(II) and Ni(II) coordination.
Inorg.Chem., 48, 2009
|
|
3DE9
 
 | | Crystal Structure of a Trimeric Cytochrome cb562 Assembly Induced by Nickel Coordination | | Descriptor: | NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | Authors: | Salgado, E.N, Lewis, R.A, Rheingold, A.L, Tezcan, F.A. | | Deposit date: | 2008-06-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through Cu(II) and Ni(II) coordination.
Inorg.Chem., 48, 2009
|
|
