4RRI
 
 | |
4RRF
 
 | | Editing domain of threonyl-tRNA synthetase from Methanococcus jannaschii with L-Ser3AA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SERINE-3'-AMINOADENOSINE, ... | | Authors: | Ahmad, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRA
 
 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Thr3AA (snapshot 1) | | Descriptor: | 3'-deoxy-3'-(L-threonylamino)adenosine, MAGNESIUM ION, Probable threonine--tRNA ligase 2 | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRB
 
 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Thr3AA (snapshot 2) | | Descriptor: | 3'-deoxy-3'-(L-threonylamino)adenosine, Probable threonine--tRNA ligase 2 | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRM
 
 | | E129A mutant of N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Thr3AA | | Descriptor: | 3'-deoxy-3'-(L-threonylamino)adenosine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ahmad, S, Muthukumar, S, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RR8
 
 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Ser3AA (snapshot 3) | | Descriptor: | MAGNESIUM ION, Probable threonine--tRNA ligase 2, SERINE-3'-AMINOADENOSINE | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRJ
 
 | |
5XYH
 
 | | Crystal Structure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae | | Descriptor: | CbsA | | Authors: | Kumar, S, Haque, A.S, Nathawat, R, Sankaranaryanan, R. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Mol. Plant Pathol., 19, 2018
|
|
5XAQ
 
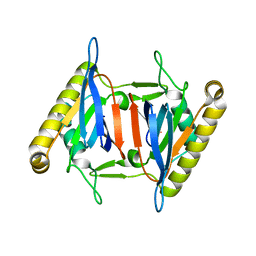 | |
1WBF
 
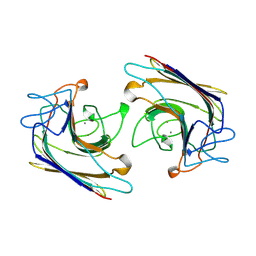 | | WINGED BEAN LECTIN, SACCHARIDE FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Suguna, K. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4Z0M
 
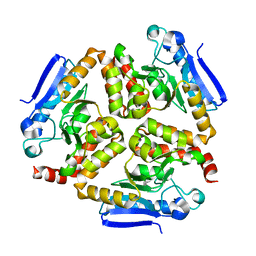 | | EchA5 Mycobacterium tuberculosis | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Chaudhary, S, Gokhale, R.S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unsaturated Lipid Assimilation by Mycobacteria Requires Auxiliary cis-trans Enoyl CoA Isomerase
Chem.Biol., 22, 2015
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4LR6
 
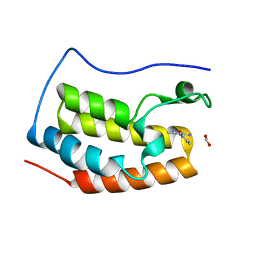 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
5YOJ
 
 | | Structure of A17 HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, A17 HIV-1 protease, GLYCEROL | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
5YOK
 
 | | Structure of HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, HIV-1 PROTEASE | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
1UGW
 
 | | Crystal structure of jacalin- Gal complex | | Descriptor: | Agglutinin alpha chain, Agglutinin alpha-chain, Agglutinin beta-3 chain, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UH1
 
 | | Crystal structure of jacalin- GalNAc-beta(1-3)-Gal-alpha-O-Me complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-methyl alpha-D-galactopyranoside, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UH0
 
 | | Crystal structure of jacalin- Me-alpha-GalNAc complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UGX
 
 | | Crystal structure of jacalin- Me-alpha-T-antigen (Gal-beta(1-3)-GalNAc-alpha-o-Me) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
3BT4
 
 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | Descriptor: | Fungal protease inhibitor-1, GLYCEROL | | Authors: | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
1M26
 
 | | Crystal structure of jacalin-T-antigen complex | | Descriptor: | Jacalin, alpha chain, beta chain, ... | | Authors: | Jeyaprakash, A.A, Rani, P.G, Reddy, G.B, Banumathi, S, Betzel, C, Surolia, A, Vijayan, M. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the jacalin-T-antigen complex and a
comparative study of lectin-T-antigen complexs
J.Mol.Biol., 321, 2002
|
|
2QXT
 
 | |
2QXU
 
 | |
1VBP
 
 | | Crystal structure of artocarpin-mannopentose complex | | Descriptor: | SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
