4FHP
 
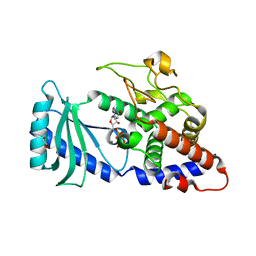 | | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity - CaUTP bound | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(A) RNA polymerase protein cid1, ... | | Authors: | Lunde, B.M, Magler, I, Meinhart, A. | | Deposit date: | 2012-06-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity.
Nucleic Acids Res., 40, 2012
|
|
1C6S
 
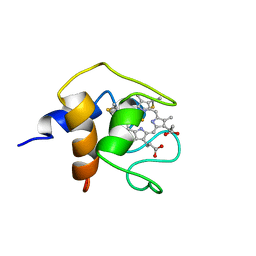 | | THE SOLUTION STRUCTURE OF CYTOCHROME C6 FROM THE THERMOPHILIC CYANOBACTERIUM SYNECHOCOCCUS ELONGATUS, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Roesch, P, Beissinger, M, Sticht, H, Sutter, M, Ejchart, A, Haehnel, W. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome c6 from the thermophilic cyanobacterium Synechococcus elongatus.
EMBO J., 17, 1998
|
|
4J32
 
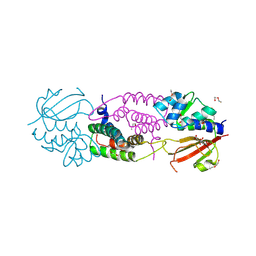 | | Structure of the effector - immunity system Tae4 / Tai4 from Salmonella typhimurium | | Descriptor: | 2-ETHOXYETHANOL, CITRATE ANION, Putative cytoplasmic protein, ... | | Authors: | Benz, J, Reinstein, J, Meinhart, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tae4/Tai4 from Salmonella typhimurium.
Plos One, 8, 2013
|
|
1Y14
 
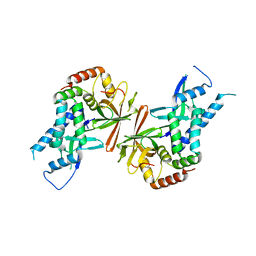 | | Crystal structure of yeast subcomplex of Rpb4 and Rpb7 | | Descriptor: | DNA-directed RNA polymerase II 19 kDa polypeptide, DNA-directed RNA polymerase II 32 kDa polypeptide | | Authors: | Armache, K.-J, Mitterweger, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Complete RNA Polymerase II and Its Subcomplex, Rpb4/7
J.Biol.Chem., 280, 2005
|
|
4J30
 
 | | Structure of the effector - immunity system Tae4 / Tai4 from Salmonella typhimurium, selenomethionine variant | | Descriptor: | 2-ETHOXYETHANOL, CITRATE ANION, Putative cytoplasmic protein, ... | | Authors: | Benz, J, Reinstein, J, Meinhart, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tae4/Tai4 from Salmonella typhimurium.
Plos One, 8, 2013
|
|
3Q8X
 
 | | Structure of a toxin-antitoxin system bound to its substrate | | Descriptor: | Antidote of epsilon-zeta postsegregational killing system, SULFATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, ... | | Authors: | Mutschler, H, Gebhardt, M, Shoeman, R.L, Meinhart, A. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis.
Plos Biol., 9, 2011
|
|
6G43
 
 | | Crystal structure of SeMet-labeled mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G44
 
 | | Crystal structure of mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | GLYCEROL, Putative major capsid protein, SULFATE ION | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G45
 
 | | Crystal structure of mavirus major capsid protein | | Descriptor: | GLYCEROL, Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G41
 
 | | Crystal structure of SeMet-labeled mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G42
 
 | | Crystal structure of mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1T9Z
 
 | | Three-dimensional structure of a RNA-polymerase II binding protein. | | Descriptor: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Kamenski, T, Heilmeier, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of
RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
2LUX
 
 | |
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
2M1I
 
 | |
3D9J
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, GLYCEROL, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9N
 
 | |
2P5T
 
 | |
3D9M
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, CTD-PEPTIDE, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9K
 
 | |
3D9I
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, GLYCEROL, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9O
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, CTD-PEPTIDE, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9L
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | ACETATE ION, CTD-PEPTIDE, GLYCEROL, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9P
 
 | |
4FH3
 
 | |
