2EZD
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZK
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, REGULARIZED MEAN STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZG
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2ODG
 
 | |
2GAT
 
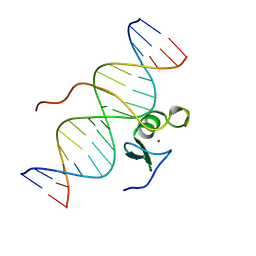 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
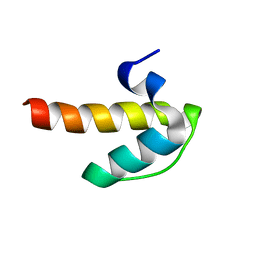
2ODC
 
 | |
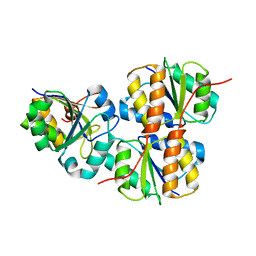
2JZN
 
 | |
2HIR
 
 | |
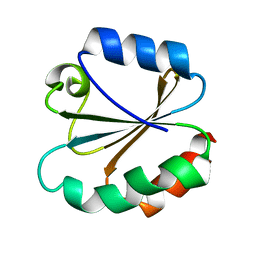
3TRX
 
 | |
4TRX
 
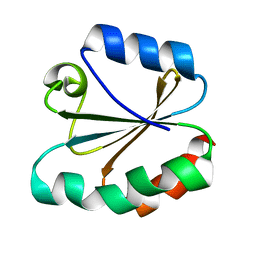 | |
1WJF
 
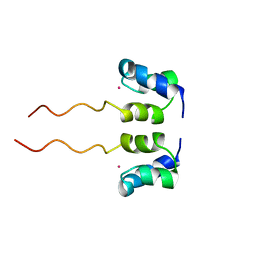 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1WCR
 
 | |
9ILB
 
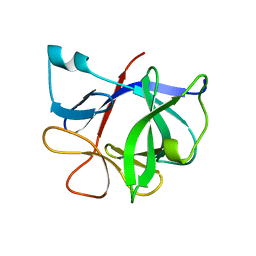 | | HUMAN INTERLEUKIN-1 BETA | | Descriptor: | PROTEIN (HUMAN INTERLEUKIN-1 BETA) | | Authors: | Yu, B, Blaber, M, Gronenborn, A.M, Clore, G.M, Caspar, D.L.D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3IL8
 
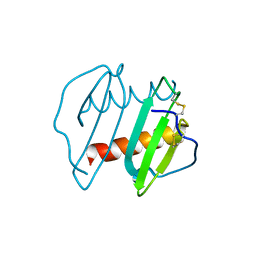 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4ZNF
 
 | |
1GB1
 
 | |
3ZNF
 
 | |
2XDF
 
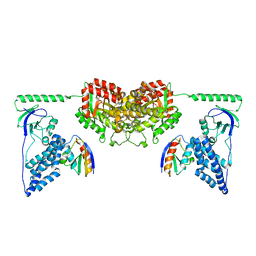 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2YHH
 
 | | Microvirin:mannobiose complex | | Descriptor: | MANNAN-BINDING LECTIN, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hussan, S, Bewley, C.A, Clore, G.M, Gustchina, E, Ghirlando, R. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Monovalent Lectin Microvirin in Complex with Man(Alpha)(1-2)Man Provides a Basis for Anti-HIV Activity with Low Toxicity.
J.Biol.Chem., 286, 2011
|
|
5I1R
 
 | | Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering.
Chemphyschem, 17, 2016
|
|
6N8C
 
 | | Structure of the Huntingtin tetramer/dimer mixture determined by paramagnetic NMR | | Descriptor: | Huntingtin | | Authors: | Schwieters, C.D, Kotler, S.A, Schmidt, T, Ceccon, A, Ghirlando, R, Libich, D.S, Clore, G.M. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1EZC
 
 | |
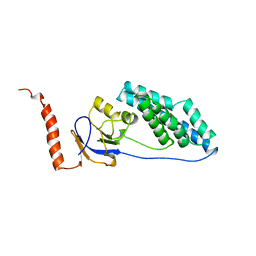
1EZA
 
 | |
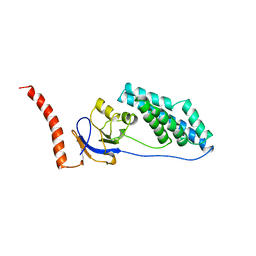
1EZB
 
 | |
6U3R
 
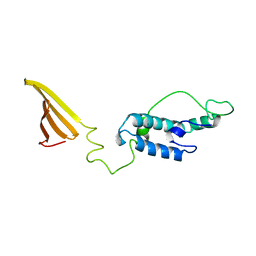 | |
