5YU1
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
1WW6
 
 | | Agrocybe cylindracea galectin complexed with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW4
 
 | | Agrocybe cylindracea galectin complexed with NeuAca2-3lactose | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-alpha-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW5
 
 | | Agrocybe cylindracea galectin complexed with 3'-sulfonyl lactose | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW7
 
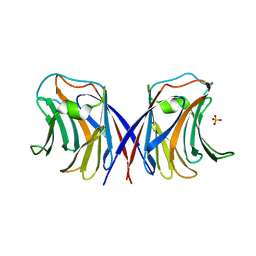 | | Agrocybe cylindracea galectin (Ligand-free) | | Descriptor: | SULFATE ION, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
7VOV
 
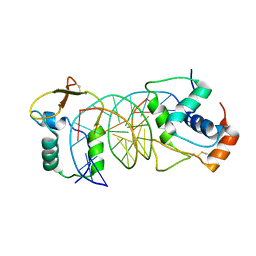 | | The crystal structure of human forkhead box protein in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOU
 
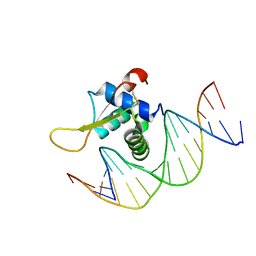 | | The crystal structure of human forkhead box protein in complex with DNA 1 | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOX
 
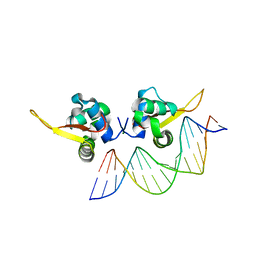 | | The crystal structure of human forkhead box protein A in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*TP*T)-3'), Hepatocyte nuclear factor 3-alpha, ... | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
4LS4
 
 | | Crystal structure of L66S mutant toxin from Helicobacter pylori | | Descriptor: | BROMIDE ION, Uncharacterized protein, Toxin | | Authors: | Pathak, C.C, Im, H, Lee, B.J, Yoon, H.J. | | Deposit date: | 2013-07-22 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4LSY
 
 | | Crystal structure of copper-bound L66S mutant toxin from Helicobacter pylori | | Descriptor: | CITRATE ANION, COPPER (II) ION, Uncharacterized protein, ... | | Authors: | Lee, B.J, Im, H, Pathak, C.C, Yoon, H.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4N9H
 
 | | Crystal structure of Transcription regulation Protein CRP | | Descriptor: | Catabolite gene activator | | Authors: | Lee, B.J, Seok, S.H, Im, H, Yoon, H.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OID
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2 | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4OIW
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4Q9D
 
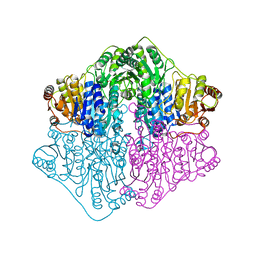 | | X-ray structure of a putative thiamin diphosphate-dependent enzyme isolated from Mycobacterium smegmatis | | Descriptor: | Benzoylformate decarboxylase, FORMIC ACID, MAGNESIUM ION | | Authors: | Andrews, F.H, Horton, J.D, Yoon, H.J, Malik, A.M.K, Logsdon, M.G, Shin, D.H, Kneen, M.M, Suh, S.W, McLeish, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The kinetic characterization and X-ray structure of a putative benzoylformate decarboxylase from M. smegmatis highlights the difficulties in the functional annotation of ThDP-dependent enzymes.
Biochim.Biophys.Acta, 1854, 2015
|
|
4Q6Q
 
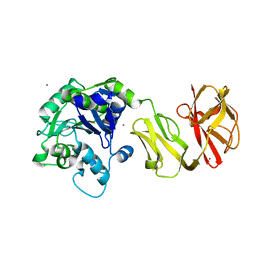 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6M
 
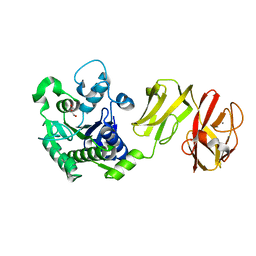 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6N
 
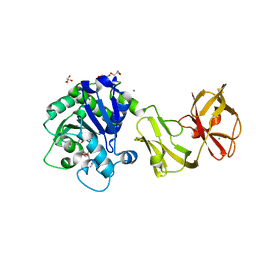 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6O
 
 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6P
 
 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QB9
 
 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | Descriptor: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | Authors: | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1HV6
 
 | | CRYSTAL STRUCTURE OF ALGINATE LYASE A1-III COMPLEXED WITH TRISACCHARIDE PRODUCT. | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-glucopyranuronic acid, ALGINATE LYASE, SULFATE ION | | Authors: | Yoon, H.-J, Hashimoto, W, Miyake, O, Murata, K, Mikami, B. | | Deposit date: | 2001-01-08 | | Release date: | 2001-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
1QAZ
 
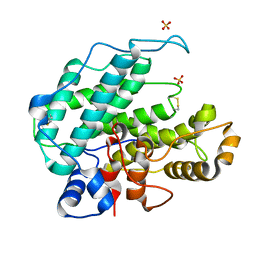 | |
4FYB
 
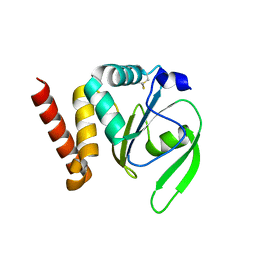 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2F5G
 
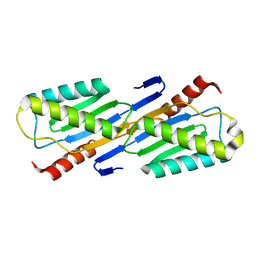 | | Crystal structure of IS200 transposase | | Descriptor: | Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2F4F
 
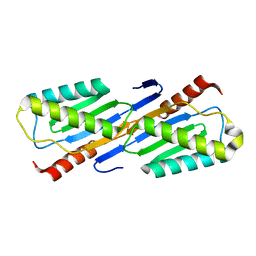 | | Crystal structure of IS200 transposase | | Descriptor: | MANGANESE (II) ION, Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
