2NV2
 
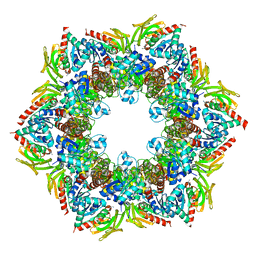 | | Structure of the PLP synthase complex Pdx1/2 (YaaD/E) from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTAMINE, ... | | Authors: | Strohmeier, M, Tews, I, Sinning, I. | | Deposit date: | 2006-11-10 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a bacterial pyridoxal 5'-phosphate synthase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2NV1
 
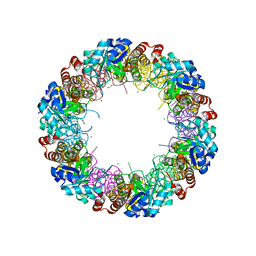 | | Structure of the synthase subunit Pdx1 (YaaD) of PLP synthase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Strohmeier, M, Tews, I, Sinning, I. | | Deposit date: | 2006-11-10 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of a bacterial pyridoxal 5'-phosphate synthase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2MVF
 
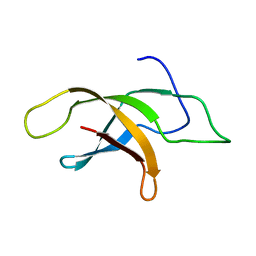 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
5NZT
 
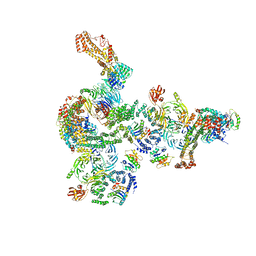 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZV
 
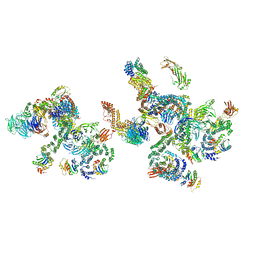 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17.299999 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZU
 
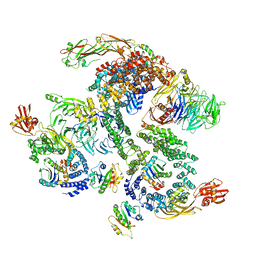 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZS
 
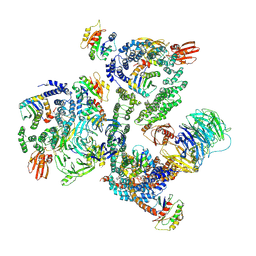 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZR
 
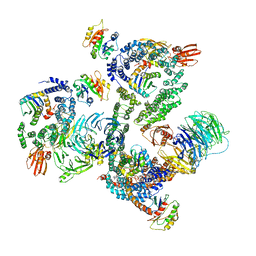 | | The structure of the COPI coat leaf | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
4GMQ
 
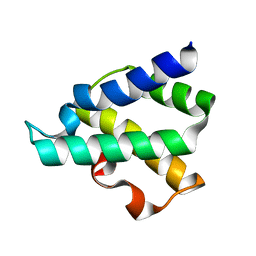 | | Ribosome-binding domain of Zuo1 | | Descriptor: | Putative ribosome associated protein | | Authors: | Kopp, J, Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GMO
 
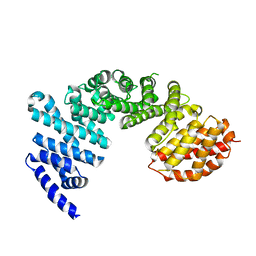 | | Crystal structure of Syo1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMN
 
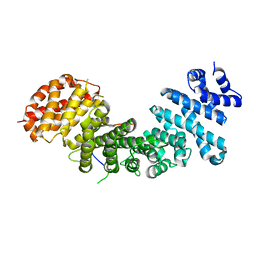 | | Structural basis of Rpl5 recognition by Syo1 | | Descriptor: | 60S ribosomal protein l5-like protein, Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GNI
 
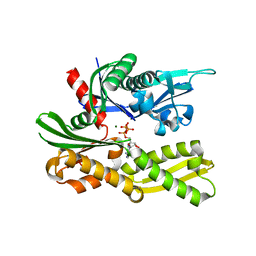 | | Structure of the Ssz1 ATPase bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3KTV
 
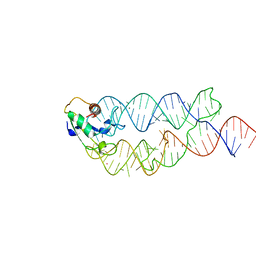 | | Crystal structure of the human SRP19/S-domain SRP RNA complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KTW
 
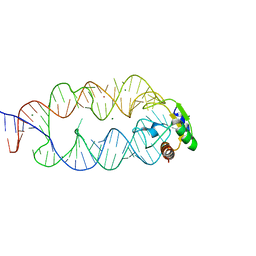 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LPZ
 
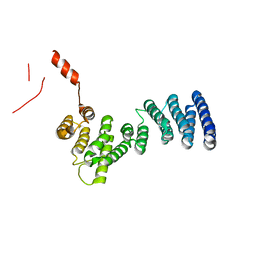 | | Crystal structure of C. therm. Get4 | | Descriptor: | Uncharacterized protein | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis.
Febs Lett., 584, 2010
|
|
3MAL
 
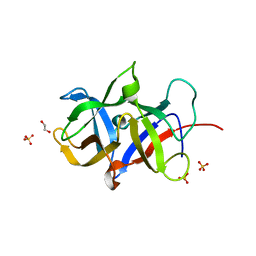 | | Crystal structure of the SDF2-like protein from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stromal cell-derived factor 2-like protein | | Authors: | Ravaud, S, Radzimanowski, J, Sinning, I. | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Arabidopsis stromal-derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum.
J.Biol.Chem., 285, 2010
|
|
3MIX
 
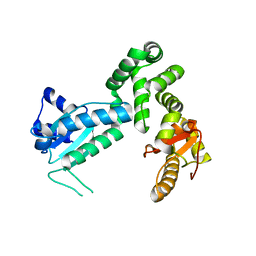 | | Crystal structure of the cytosolic domain of B. subtilis FlhA | | Descriptor: | Flagellar biosynthesis protein flhA | | Authors: | Bange, G, Kuemmerer, N, Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DEP
 
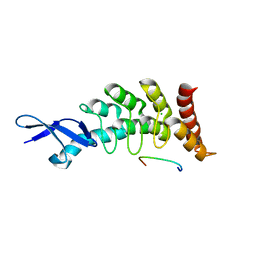 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
3SJC
 
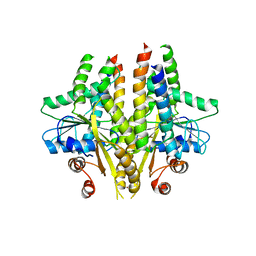 | |
3SYN
 
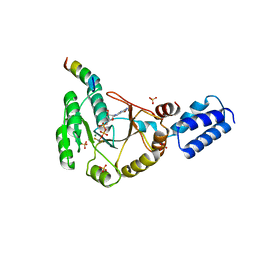 | | Crystal structure of FlhF in complex with its activator | | Descriptor: | ALUMINUM FLUORIDE, ATP-binding protein YlxH, Flagellar biosynthesis protein flhF, ... | | Authors: | Bange, G, Kuemmerer, N, Wild, K, Sinning, I. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Structural basis for the molecular evolution of SRP-GTPase activation by protein.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SJA
 
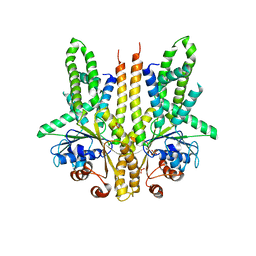 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3ZRI
 
 | |
3ZRJ
 
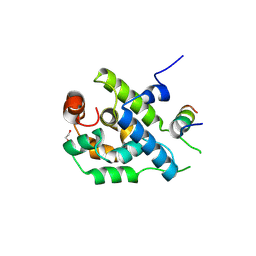 | | Complex of ClpV N-domain with VipB peptide | | Descriptor: | 1,2-ETHANEDIOL, CLPB PROTEIN, VIPB | | Authors: | Lenherr, E.D, Kopp, J, Sinning, I. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Basis for the Unique Role of the Aaa+ Chaperone Clpv in Type Vi Protein Secretion.
J.Biol.Chem., 286, 2011
|
|
3SJD
 
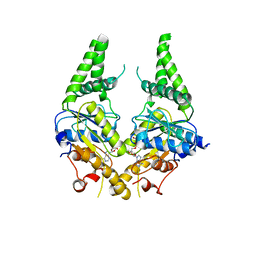 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
4B9Q
 
 | |
