1HQ2
 
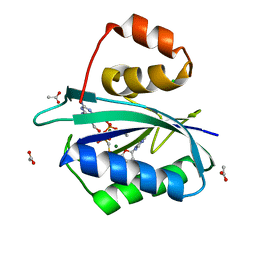 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF E.COLI HPPK(R82A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.25 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ACETATE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-12-13 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
4JBA
 
 | |
1G4C
 
 | |
4QOC
 
 | |
8WRH
 
 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRM
 
 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
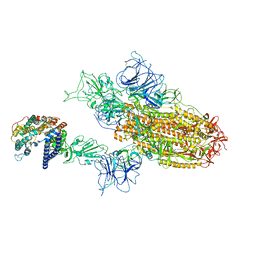 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
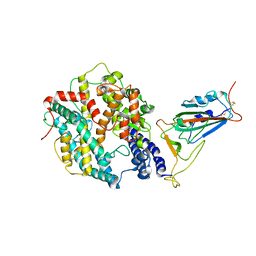 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
5ICA
 
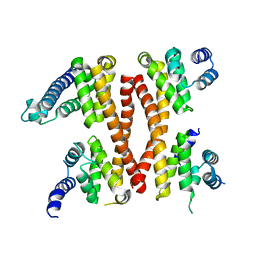 | | Structure of the CTD complex of UTP12, Utp13, Utp1 and Utp21 | | Descriptor: | Periodic tryptophan protein 2-like protein, Putative U3 snoRNP protein, Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.507 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC7
 
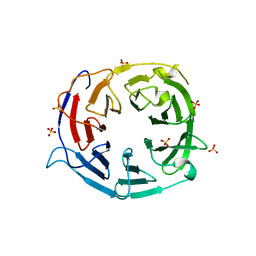 | | Structure of the WD domain of UTP18 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.331 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC9
 
 | | Structure of the CTD complex of Utp12 and Utp13 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC8
 
 | | Structure of UTP6 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
1F9H
 
 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF E. COLI HPPK(R92A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.50 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-07-10 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
4A6L
 
 | | beta-tryptase inhibitor | | Descriptor: | 1-{3-[1-({5-[(2-fluorophenyl)ethynyl]furan-2-yl}carbonyl)piperidin-4-yl]phenyl}methanamine, TRYPTASE ALPHA/BETA-1 | | Authors: | Mathieu, M, Maignan, S. | | Deposit date: | 2011-11-04 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Library Design and the Discovery of a Potent and Selective Mast Cell Beta-Tryptase Inhibitor as an Oral Therapeutic Agent.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1IRB
 
 | | CARBOXYLIC ESTER HYDROLASE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-08-13 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phospholipase A2 engineering. Deletion of the C-terminus segment changes substrate specificity and uncouples calcium and substrate binding at the zwitterionic interface.
Biochemistry, 35, 1996
|
|
6X3K
 
 | |
6X3Q
 
 | |
6XTG
 
 | | Ab 1116NS19.9 bound to CA19-9 | | Descriptor: | Heavy chain, Light chain, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUN
 
 | | Ab 5b1 bound to CA19-9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUK
 
 | | AbLIFT design 15 of Ab 1116NS19.9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUD
 
 | | Apo Ab 1116NS19.9 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUL
 
 | | Apo Ab 5b1 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
4LA3
 
 | |
