6UIN
 
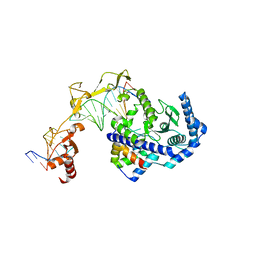 | | Role of Beta-hairpin motifs in the DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*NP*NP*NP*NP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | Tethering-facilitated DNA 'opening' and complementary roles of beta-hairpin motifs in the Rad4/XPC DNA damage sensor protein
Nucleic Acids Res., 48, 2021
|
|
8H6T
 
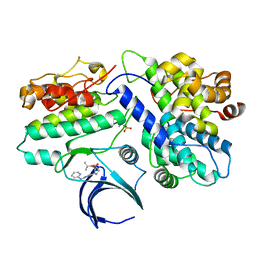 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6P
 
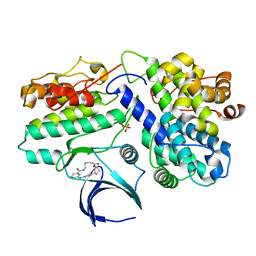 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8KB0
 
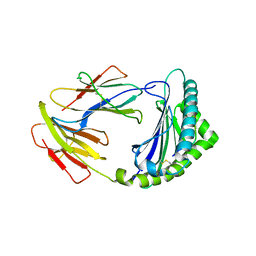 | | Crystal structure of 01JD-AEAIIVAMV | | Descriptor: | Beta2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KB1
 
 | | Crystal structure of 11JD | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
4IGQ
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 in complex with methylated H3K4 substrate | | Descriptor: | FE (III) ION, N-OXALYLGLYCINE, Os05g0196500 protein, ... | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
6CDI
 
 | | Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDM
 
 | |
4IGP
 
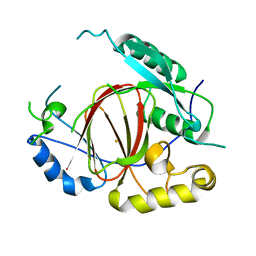 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 apo enzyme | | Descriptor: | FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGO
 
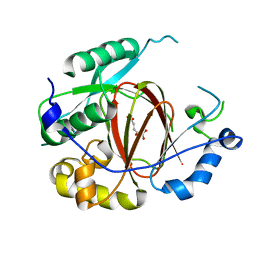 | | Histone H3 Lysine 4 Demethylating rice Rice JMJ703 in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDO
 
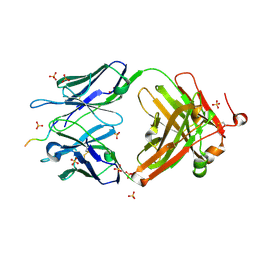 | | Structure of vaccine-elicited HIV-1 neutralizing antibody vFP16.02 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP16.02 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDP
 
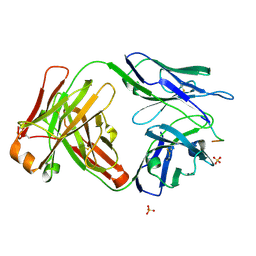 | | Vaccine-elicited HIV-1 neutralizing antibody vFP20.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP20.01 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
4IU6
 
 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | Descriptor: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | Deposit date: | 2013-01-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
1HQ2
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF E.COLI HPPK(R82A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.25 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ACETATE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-12-13 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
7Y56
 
 | |
7Y57
 
 | |
7YEP
 
 | | 2-APB bound state of mTRPV2 | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
1KQ8
 
 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1Q0N
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE FROM E. COLI WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.25 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Catalytic Center Assembly of Hppk as Revealed by the Crystal Structure of a Ternary Complex at 1.25 A Resolution
Structure, 8, 2000
|
|
7R0Q
 
 | | KRasG12C in complex with GDP and compound 3 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0M
 
 | | KRasG12C in complex with GDP and JDQ443 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-(1-methylindazol-5-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
6EFC
 
 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFI
 
 | | SK678 binding region (Siglec + Unique) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
