3FCY
 
 | |
5ABS
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL COILED-COIL DOMAIN OF CIN85 IN SPACE GROUP P321 | | Descriptor: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1, ZINC ION | | Authors: | Wong, L, Habeck, M, Griesinger, C, Becker, S. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-13 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Adaptor Protein Cin85 Assembles Intracellular Signaling Clusters for B Cell Activation.
Sci.Signal., 9, 2016
|
|
7PL9
 
 | |
6NI4
 
 | |
6NI7
 
 | |
6NPQ
 
 | |
5HI8
 
 | | Structure of T-type Phycobiliprotein Lyase CpeT from Prochlorococcus phage P-HM1 | | Descriptor: | ACETATE ION, Antenna protein, MAGNESIUM ION | | Authors: | Gasper, R, Schwach, J, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct Features of Cyanophage-encoded T-type Phycobiliprotein Lyase Phi CpeT: THE ROLE OF AUXILIARY METABOLIC GENES.
J. Biol. Chem., 292, 2017
|
|
6NIA
 
 | |
6NI9
 
 | |
6NI5
 
 | |
6NI6
 
 | |
5HG7
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-{(3R,4R)-3-[5-Chloro-2-(1-methyl-1H-pyrazol-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-yloxymethyl]-4-methoxy-pyrrolidin-1-yl}propenone (PF-06459988) | | Descriptor: | 1-{(3R,4R)-3-[({5-chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}propan-1-one, Epidermal growth factor receptor, SULFATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG5
 
 | | EGFR (L858R, T790M, V948R) in complex with N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}propanamide, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG9
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | Descriptor: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG8
 
 | | EGFR (L858R, T790M, V948R) in complex with N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]propanamide, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
3DKS
 
 | | DsbA substrate complex | | Descriptor: | Thiol:disulfide interchange protein dsbA, siga peptide | | Authors: | Paxman, J.J, Borg, N.A, Horne, J, Rossjohn, J, Thompson, P.E, Piek, S, Kahler, C.M, Sakellaris, H, Scanlon, M.J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
J.Biol.Chem., 284, 2009
|
|
2A4A
 
 | | Deoxyribose-phosphate aldolase from P. yoelii | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Walker, J.R, Amani, M, Lew, J, Wiegelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
7KJS
 
 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | Descriptor: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | McTigue, M.A, He, Y, Ferre, R.A. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6UND
 
 | |
6UNF
 
 | |
8V5W
 
 | | UIC-1 mutant UIC-1-B5T | | Descriptor: | UIC-1-B5T | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V61
 
 | | UIC-1 mutant - UIC-1-L6I | | Descriptor: | UIC-1-L6I | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5X
 
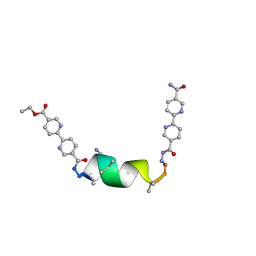 | | UIC-1 mutant - UIC-1-L6A | | Descriptor: | ACETONITRILE, UIC-1-L6A | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V59
 
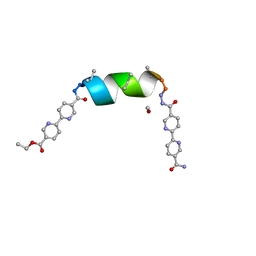 | | UIC-1 mutant - UIC-1-B5I | | Descriptor: | METHANOL, UIC-1-B5I | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-11-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5Z
 
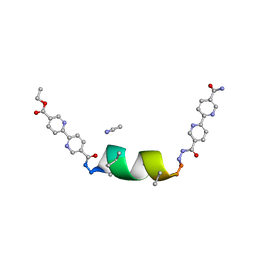 | | UIC-1 mutant - UIC-1-L6M | | Descriptor: | ACETONITRILE, UIC-1-L6M | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
