6MW3
 
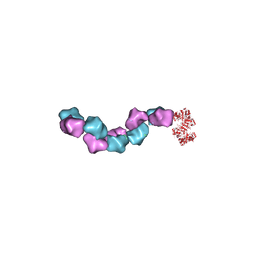 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4N83
 
 | |
4ERM
 
 | |
4ERP
 
 | |
5CLG
 
 | |
5CLJ
 
 | |
1AV8
 
 | | RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT FROM E. COLI | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
5CI4
 
 | | Ribonucleotide reductase beta subunit | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI3
 
 | | Ribonucleotide reductase Y122 2,3,5-F3Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 subunit beta, SULFATE ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI2
 
 | | Ribonucleotide reductase Y122 2,3,6-F3Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 subunit beta, SULFATE ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI1
 
 | | Ribonucleotide reductase Y122 2,3-F2Y variant | | Descriptor: | CHLORIDE ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI0
 
 | | Ribonucleotide reductase Y122 3,5-F2Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
2NSL
 
 | | E. coli PurE H45N mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2NSH
 
 | | E. coli PurE H45Q mutant complexed with nitro-AIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2NSJ
 
 | | E. coli PurE H45Q mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2HS0
 
 | | T. maritima PurL complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HRY
 
 | | T. maritima PurL complexed with AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HRU
 
 | | T. maritima PurL complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS3
 
 | | T. maritima PurL complexed with FGAR | | Descriptor: | N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS4
 
 | | T. maritima PurL complexed with FGAR and AMPPCP | | Descriptor: | MAGNESIUM ION, N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
8RK1
 
 | | Crystal structure of FutA bound to Fe(III) solved by neutron diffraction | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-12-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (2.095 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8C4Y
 
 | | SFX structure of FutA bound to Fe(III) | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-01-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OEI
 
 | | SFX structure of FutA after an accumulated dose of 350 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OEM
 
 | | Crystal structure of FutA bound to Fe(II) | | Descriptor: | FE (II) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OGG
 
 | | Crystal structure of FutA after an accumulated dose of 5 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
