6JKR
 
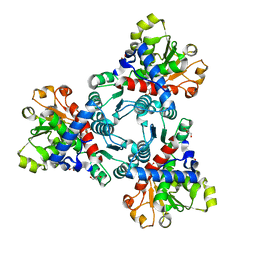 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl phosphate (CP) | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL4
 
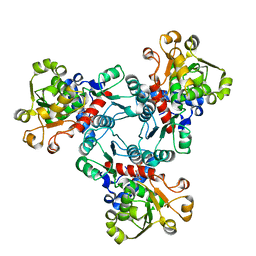 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl aspartate (CA) and phosphate (Pi) | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JKS
 
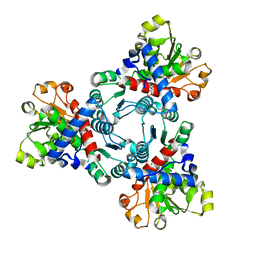 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl phosphate (CP) and aspartate (Asp) | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, putative, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL5
 
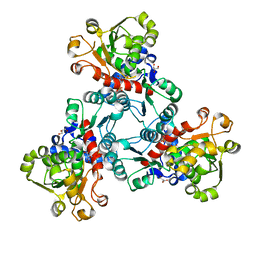 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with aspartate (Asp) and phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
1WR6
 
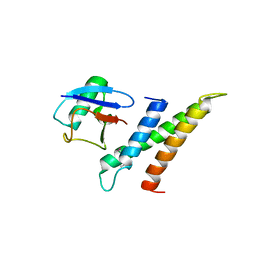 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
7E3N
 
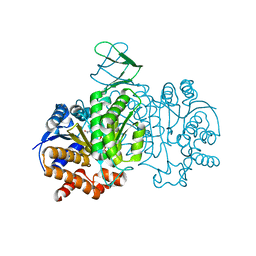 | | Crystal structure of Isocitrate dehydrogenase D252N mutant from Trypanosoma brucei in complexed with NADP+, alpha-ketoglutarate, and ca2+ | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], ... | | Authors: | Arai, N, Shiba, T, Inaoka, D.K, Kita, K, Wang, X, Otani, M, Matsushiro, S, Kojima, C. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Isocitrate dehydrogenase from Trypanosoma brucei.
To Be Published
|
|
6J9Q
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with AMP-PNP. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
6J9V
 
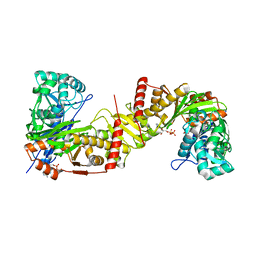 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
5YJX
 
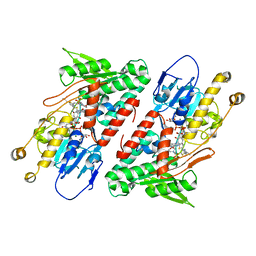 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with myxothiazol. | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJY
 
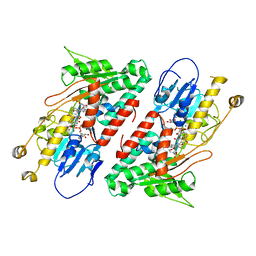 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with AC0-12. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-dodecyl-1-oxidanidyl-quinolin-1-ium-4-ol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJW
 
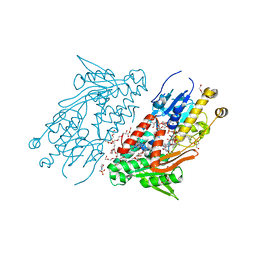 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with the competitive inhibitor, stigmatellin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5ZFB
 
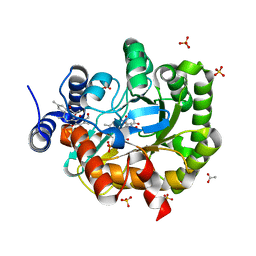 | | Structure of human dihydroorotate dehydrogenase in complex with ascofuranone (open-form) | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2Z,6Z,8E)-11-hydroxy-3,7,11-trimethyl-10-oxododeca-2,6,8-trien-1-yl]-2-methylbenzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
5ZF4
 
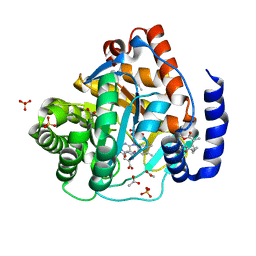 | | Structure of human dihydroorotate dehydrogenase in complex with 275-10-COOMe | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
5ZFA
 
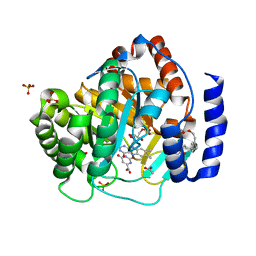 | | Structure of human dihydroorotate dehydrogenase in complex with 287-12-OCOiPr | | Descriptor: | (2E,6E)-8-(3-chloro-5-formyl-2,6-dihydroxy-4-methylphenyl)-3,6-dimethylocta-2,6-dien-1-yl 2-methylpropanoate, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
5ZF8
 
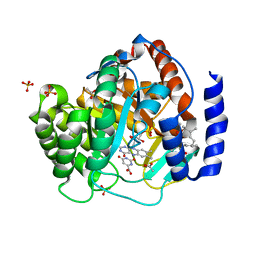 | | Structure of human dihydroorotate dehydrogenase in complex with 277-11-OAc | | Descriptor: | (2S,3E,7E)-9-(3-chloro-5-formyl-2,6-dihydroxy-4-methylphenyl)-3,7-dimethylnona-3,7-dien-2-yl acetate, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
5ZF7
 
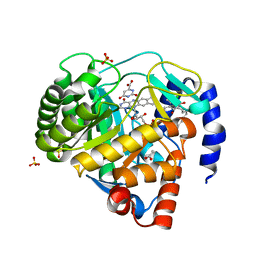 | | Structure of human dihydroorotate dehydrogenase in complex with 277-9-OH | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2E,6E,8S)-8-hydroxy-3,7-dimethylnona-2,6-dien-1-yl]-2-methylbenzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
5ZF9
 
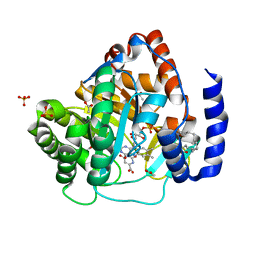 | | Structure of human dihydroorotate dehydrogenase in complex with 280-12 | | Descriptor: | 3-chloro-4,6-dihydroxy-2-methyl-5-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]benzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
3AGR
 
 | |
2ZYZ
 
 | | Pyrobaculum aerophilum splicing endonuclease | | Descriptor: | Putative uncharacterized protein PAE0789, tRNA-splicing endonuclease | | Authors: | Yoshinari, S, Inaoka, D.K, Watanabe, Y, Shiba, T, Kurisu, G, Harada, S. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease
Nucleic Acids Res., 37, 2009
|
|
3AEQ
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AES
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AEK
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AET
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AEU
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AER
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
