4ZK5
 
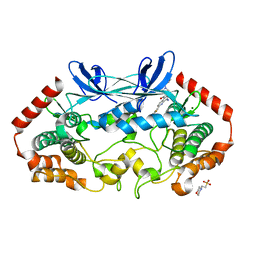 | | MAP4K4 in complex with inhibitor GNE-495 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-amino-N-[1-(cyclopropylcarbonyl)azetidin-3-yl]-2-(3-fluorophenyl)-1,7-naphthyridine-5-carboxamide, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2015-04-29 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Based Design of GNE-495, a Potent and Selective MAP4K4 Inhibitor with Efficacy in Retinal Angiogenesis.
Acs Med.Chem.Lett., 6, 2015
|
|
6GY9
 
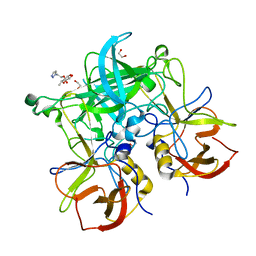 | | Fucose-functionalized precision glycomacromolecules targeting human norovirus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,3-triazol-1-yl)ethyl 6-deoxy-alpha-L-galactopyranoside, Capsid protein | | Authors: | Ruoff, K, Kilic, T, Hansman, G.S. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fucose-Functionalized Precision Glycomacromolecules Targeting Human Norovirus Capsid Protein.
Biomacromolecules, 19, 2018
|
|
1LBM
 
 | |
5HVY
 
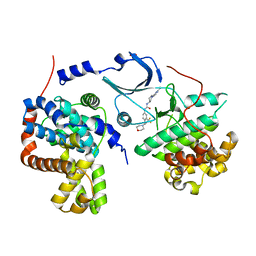 | | CDK8/CYCC IN COMPLEX WITH COMPOUND 20 | | Descriptor: | CHLORIDE ION, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Bergeron, P, Koehler, M. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Development of a Series of Potent and Selective Type II Inhibitors of CDK8.
Acs Med.Chem.Lett., 7, 2016
|
|
4QYE
 
 | | CHK1 kinase domain in complex with diarylpyrazine compound 1 | | Descriptor: | 4-[6-(3-hydroxyphenyl)pyrazin-2-yl]benzoic acid, Serine/threonine-protein kinase Chk1 | | Authors: | Appleton, B, Wu, P. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of the 1,7-diazacarbazole class of inhibitors of checkpoint kinase 1.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7ZR2
 
 | | Crystal structure of a chimeric protein mimic of SARS-CoV-2 Spike HR1 in complex with HR2 | | Descriptor: | Spike protein S2', Spike protein S2',Chimeric protein mimic of SARS-CoV-2 Spike HR1 | | Authors: | Camara-Artigas, A, Gavira, J.A, Cano-Munoz, M, Polo-Megias, D, Conejero-Lara, F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel chimeric proteins mimicking SARS-CoV-2 spike epitopes with broad inhibitory activity.
Int.J.Biol.Macromol., 222, 2022
|
|
4C1A
 
 | |
2YHB
 
 | |
2YKP
 
 | |
2YKQ
 
 | |
2YHA
 
 | |
4C1B
 
 | |
2YKO
 
 | |
5CEI
 
 | | Crystal structure of CDK8:Cyclin C complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-iodophenoxy)-N-methylthieno[2,3-c]pyridine-2-carboxamide, Cyclin-C, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Koehler, M.F.T. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Development of a Potent, Specific CDK8 Kinase Inhibitor Which Phenocopies CDK8/19 Knockout Cells.
Acs Med.Chem.Lett., 7, 2016
|
|
4U3Y
 
 | |
4U41
 
 | | MAP4K4 Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U45
 
 | | MAP4K4 in complex with inhibitor (compound 25) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(1H-pyrazol-4-yl)-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U3Z
 
 | | APO MAP4K4 T181E Phosphomimetic Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase kinase kinase kinase 4, SODIUM ION | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U43
 
 | | MAP4K4 in complex with inhibitor (compound 6) | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 4, N-(pyridin-3-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U40
 
 | |
4U42
 
 | | MAP4K4 T181E Mutant Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U44
 
 | | MAP4K4 in complex with inhibitor (compound 16) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-phenyl-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6HQG
 
 | | Cytochrome P450-153 from Phenylobacterium zucineum | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQW
 
 | |
