4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4WW1
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8 and Beta Chain-TRBV7-8 | | Descriptor: | TCR Alpha Chain-TRAV21-TRAJ8, TCR Beta Chain-TRBV7-8 | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Lim, R.T, Besra, G, Howell, A.R, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
3QZ0
 
 | | Structure of Treponema denticola Factor H Binding protein (FhbB), selenomethionine derivative | | Descriptor: | Factor H binding protein, GLYCEROL, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Heroux, A, Bell, J.K, Marconi, R.T, Conrad, D.H, Burgner, J.W. | | Deposit date: | 2011-03-04 | | Release date: | 2012-03-07 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of factor H-binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into progression of periodontal disease.
J.Biol.Chem., 287, 2012
|
|
3R15
 
 | | Structure Treponema Denticola Factor H Binding Protein | | Descriptor: | Factor H binding protein, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Burgner, J, Heroux, A, Bell, J.K, Marconi, R.T. | | Deposit date: | 2011-03-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure Treponema Denticola Factor H Binding Protein
To be Published
|
|
4LVW
 
 | |
7UIX
 
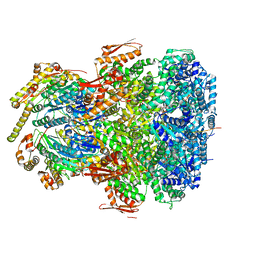 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIY
 
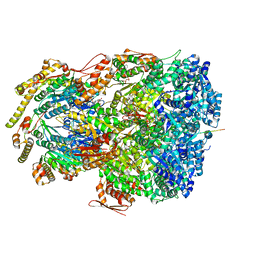 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
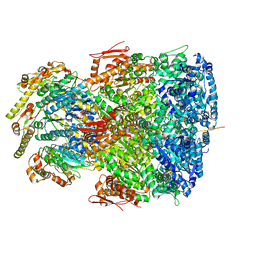 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
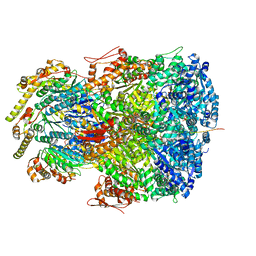 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4LVX
 
 | |
4LVZ
 
 | |
4LVY
 
 | | Structure of the THF riboswitch bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LVV
 
 | | Structure of the THF riboswitch | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LW0
 
 | | Structure of the THF riboswitch bound to adenine | | Descriptor: | ADENINE, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
8S98
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 8 | | Descriptor: | (8S)-N-cyclopropyl-5-[(2-methoxypyridin-3-yl)amino]-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S9A
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with TAK-279 | | Descriptor: | (8S)-N-[(1R,2R)-2-methoxycyclobutyl]-7-(methylamino)-5-{[(1P,2'P)-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S99
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 11 | | Descriptor: | (8S)-N-[(1R,2S)-2-fluorocyclopropyl]-5-{[(1M,2'M)-3'-fluoro-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8SMU
 
 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
4BFA
 
 | | Crystal structure of E. coli dihydrouridine synthase C (DusC) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NITRATE ION, ... | | Authors: | Byrne, R.T, Whelan, F, Konevega, A, Aziz, N, Rodnina, M, Antson, A.A. | | Deposit date: | 2013-03-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Major Reorientation of tRNA Substrates Defines Specificity of Dihydrouridine Synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4BF9
 
 | | Crystal structure of E. coli dihydrouridine synthase C (DusC) (selenomethionine derivative) | | Descriptor: | FLAVIN MONONUCLEOTIDE, TRNA-DIHYDROURIDINE SYNTHASE C | | Authors: | Byrne, R.T, Whelan, F, Konevega, A, Aziz, N, Rodnina, M, Antson, A.A. | | Deposit date: | 2013-03-16 | | Release date: | 2013-11-06 | | Last modified: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Major Reorientation of tRNA Substrates Defines Specificity of Dihydrouridine Synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8SQX
 
 | |
8U9O
 
 | |
4ERL
 
 | |
