7YQ7
 
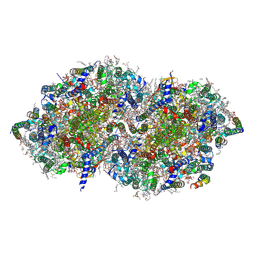 | | Crystal structure of photosystem II expressing psbA3 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
8HX6
 
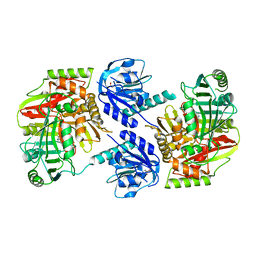 | |
8HX7
 
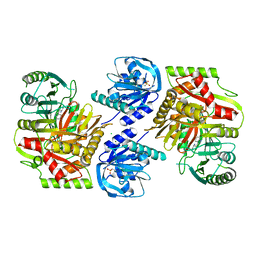 | |
8HX9
 
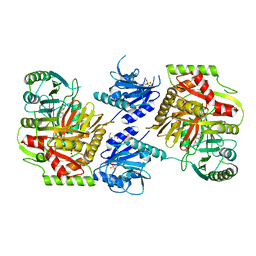 | | Crystal structure of 4-amino-4-deoxychorismate synthase from Streptomyces venezuelae with chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-amino-4-deoxychorismate synthase, FORMIC ACID, ... | | Authors: | Nakamichi, Y, Watanabe, M. | | Deposit date: | 2023-01-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the allosteric pathway of 4-amino-4-deoxychorismate synthase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8HX8
 
 | |
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
2Z2E
 
 | | Crystal Structure of Canine Milk Lysozyme Stabilized against Non-enzymatic Deamidation | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Nonaka, Y, Akieda, D, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Spontaneous asparaginyl deamidation of canine milk lysozyme under mild conditions
Proteins, 72, 2008
|
|
1WSF
 
 | | Co-crystal structure of E.coli RNase HI active site mutant (D134A*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
1WSE
 
 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
1WSG
 
 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A/D134N*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
7YBC
 
 | |
7YB8
 
 | |
7YBB
 
 | |
7YBA
 
 | |
7YB9
 
 | |
1IS2
 
 | |
1X0J
 
 | | Crystal structure analysis of the N-terminal bromodomain of human Brd2 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-23 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for acetylated histone H4 recognition by the human Brd2 bromodomain
To be Published
|
|
1V93
 
 | | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5,10-Methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakajima, Y, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of 5,10-Methylenetetrahydrofolate reductase from Thermus thermophilus HB8
To be Published
|
|
7VKF
 
 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y, Nishiya, Y, Ito, K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VKD
 
 | |
7VZS
 
 | | FAD-dpendent Glucose Dehydrogenase complexed with an inhibitor at pH7.56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-glucal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VZP
 
 | | FAD-dpendent Glucose Dehydrogenase from Aspergillus oryzae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase, PENTAETHYLENE GLYCOL | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
2ECF
 
 | |
2DZO
 
 | |
