6U8S
 
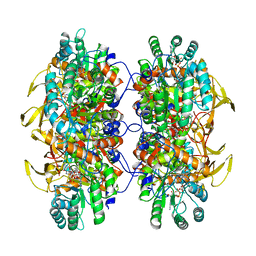 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Filament assembly interface reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UA2
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (2/4 compressed, 2/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UAJ
 
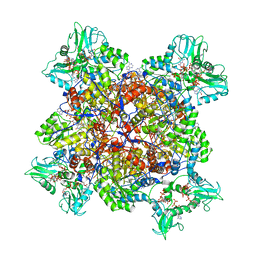 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free canonical octamer reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UC2
 
 | |
7RES
 
 | | HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFI
 
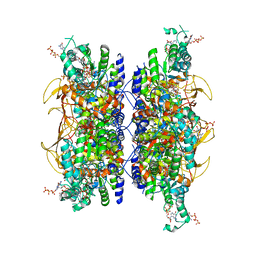 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFG
 
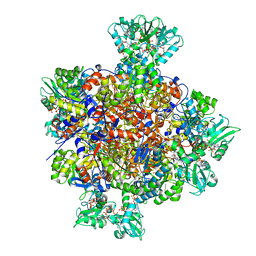 | | HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+ OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFE
 
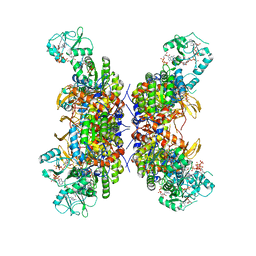 | | HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+; INTERFACE-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFF
 
 | |
7RGD
 
 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5FM1
 
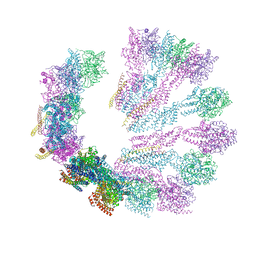 | | Structure of gamma-tubulin small complex based on a cryo-EM map, chemical cross-links, and a remotely related structure | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-30 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
5FLZ
 
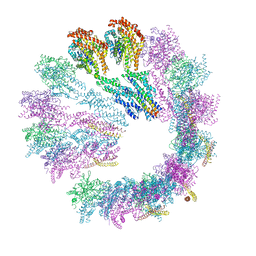 | | Cryo-EM structure of gamma-TuSC oligomers in a closed conformation | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
8G9J
 
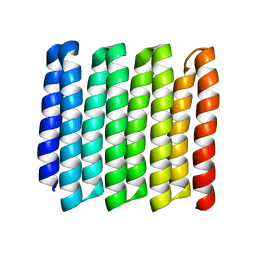 | |
8G9K
 
 | |
8GA6
 
 | |
8GA7
 
 | |
8GEL
 
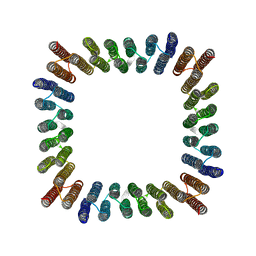 | | Cryo-EM structure of synthetic tetrameric building block sC4 | | Descriptor: | sC4 | | Authors: | Redler, R.L, Huddy, T.F, Hsia, Y, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Blueprinting extendable nanomaterials with standardized protein blocks.
Nature, 627, 2024
|
|
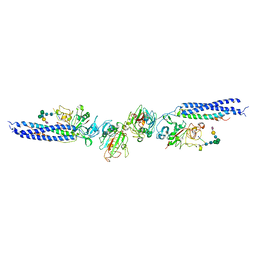
3H32
 
 | |
3RIP
 
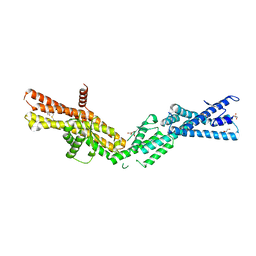 | | Crystal Structure of human gamma-tubulin complex protein 4 (GCP4) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, Gamma-tubulin complex component 4 | | Authors: | Gregory-Pauron, L, Guillet, V, Mourey, L. | | Deposit date: | 2011-04-14 | | Release date: | 2011-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of gamma-tubulin complex protein GCP4 provides insight into microtubule nucleation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8UBG
 
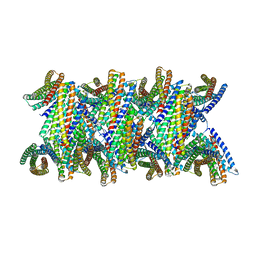 | | DpHF19 filament | | Descriptor: | DpHF19,Green fluorescent protein (Fragment) | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8UAO
 
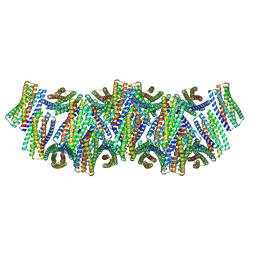 | | DpHF18 filament | | Descriptor: | DpHF18 | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-21 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8UB3
 
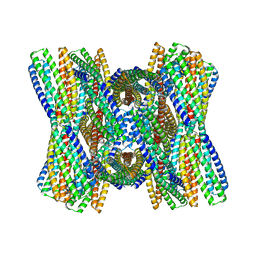 | | DpHF7 filament | | Descriptor: | DpHF7 filament | | Authors: | Lynch, E.M, Farrell, D, Shen, H, Kollman, J.M, DiMaio, F, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
6XQW
 
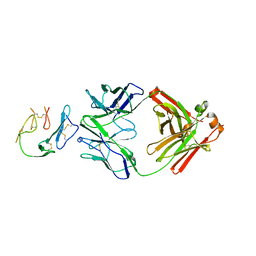 | | Crystal Structure of MaliM03 Fab in complex with Pfmsp1-19 | | Descriptor: | MaliM03 Fab Heavy Chain, MaliM03 Fab Light Chain, Pfmsp1-19 | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Multimeric antibodies from antigen-specific human IgM+ memory B cells restrict Plasmodium parasites.
J.Exp.Med., 218, 2021
|
|
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6P4X
 
 | |
