2O4C
 
 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
3LDI
 
 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | GLYCEROL, MERCURY (II) ION, Pancreatic trypsin inhibitor, ... | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3LDJ
 
 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, Pancreatic trypsin inhibitor | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding.
Biochem.Biophys.Res.Commun., 2010
|
|
3LDM
 
 | |
3JTR
 
 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JTQ
 
 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3OJE
 
 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase (Apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3OJF
 
 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase with NADP+ and indole naphthyridinone (Complex form) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
6C70
 
 | |
2R64
 
 | | Crystal structure of a 3-aminoindazole compound with CDK2 | | Descriptor: | Cell division protein kinase 2, N-[5-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-1H-INDAZOL-3-YL]-2-(4-PIPERIDIN-1-YLPHENYL)ACETAMIDE | | Authors: | Lee, J, Choi, H, Kim, K.H, Jeong, S, Park, J.W, Baek, C.S, Lee, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and biological evaluation of 3,5-diaminoindazoles as cyclin-dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3R4Y
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3QWZ
 
 | | Crystal structure of FAF1 UBX-p97N-domain complex | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-02-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
3QX1
 
 | | Crystal structure of FAF1 UBX domain | | Descriptor: | FAS-associated factor 1, SULFATE ION | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
6ILQ
 
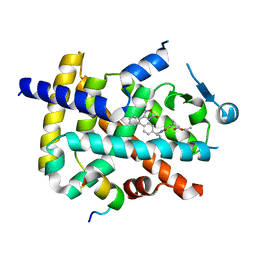 | | Crystal structure of PPARgamma with compound BR101549 | | Descriptor: | Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma, ethyl [2-butyl-6-oxo-1-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}-4-(propan-2-yl)-1,6-dihydropyrimidin-5-yl]acetate | | Authors: | Hong, E, Jang, T.H, Chin, J, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-10-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Identification of BR101549 as a lead candidate of non-TZD PPAR gamma agonist for the treatment of type 2 diabetes: Proof-of-concept evaluation and SAR.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3S8R
 
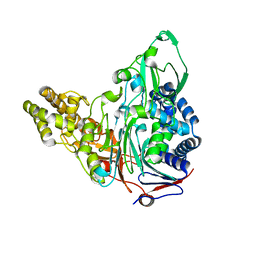 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | GLYCEROL, Glutaryl-7-aminocephalosporanic-acid acylase | | Authors: | Kim, J.K, Yang, I.S, Park, S.S, Kim, K.H. | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation.
Biochemistry, 42, 2003
|
|
1OR0
 
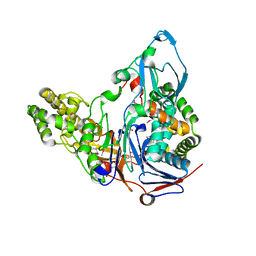 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | 1,2-ETHANEDIOL, Glutaryl 7-Aminocephalosporanic Acid Acylase, glutaryl acylase | | Authors: | Kim, J.K, Yang, I.S, Rhee, S, Dauter, Z, Lee, Y.S, Park, S.S, Kim, K.H. | | Deposit date: | 2003-03-11 | | Release date: | 2004-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation
Biochemistry, 42, 2003
|
|
1VDD
 
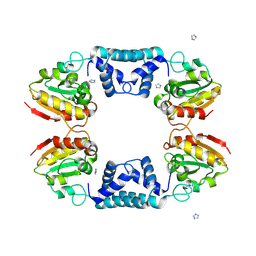 | |
1R5G
 
 | | Crystal Structure of MetAP2 complexed with A311263 | | Descriptor: | (2S,3R)-3-AMINO-2-HYDROXY-5-(ETHYLSULFANYL)PENTANOYL-((S)-(-)-(1-NAPHTHYL)ETHYL)AMIDE, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1RUC
 
 | | RHINOVIRUS 14 MUTANT N1105S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUI
 
 | | RHINOVIRUS 14 MUTANT S1223G COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUG
 
 | | RHINOVIRUS 14 MUTANT N1219S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUF
 
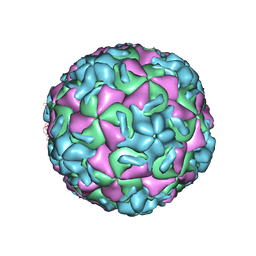 | | RHINOVIRUS 14 (HRV14) (MUTANT WITH ASN 1 219 REPLACED BY ALA (N219A IN CHAIN 1) | | Descriptor: | RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUJ
 
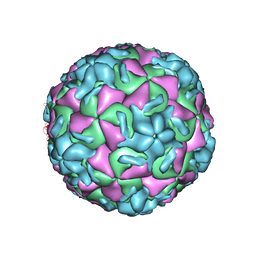 | | RHINOVIRUS 14 MUTANT WITH SER 1 223 REPLACED BY GLY (S1223G) | | Descriptor: | RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUE
 
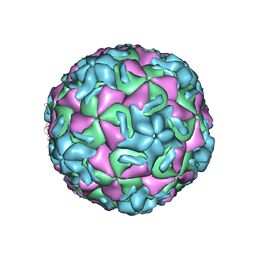 | | RHINOVIRUS 14 SITE DIRECTED MUTANT N1219A COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
