6HPV
 
 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
1BL6
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1A9U
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3KMW
 
 | | Crystal structure of the ILK/alpha-parvin core complex (MgATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-parvin, Integrin-linked kinase, ... | | Authors: | Fukuda, K, Qin, J. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions.
Mol.Cell, 36, 2009
|
|
3KMU
 
 | |
7ND4
 
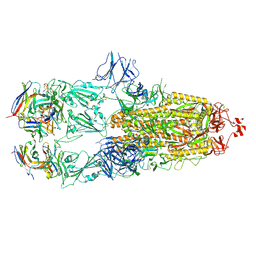 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-88 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND3
 
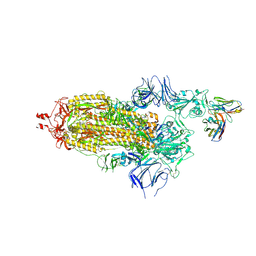 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-40 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND8
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-384 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND7
 
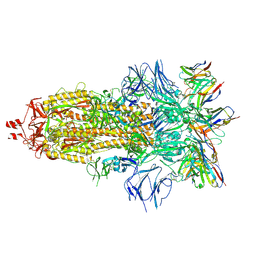 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDC
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND9
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-253H55L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDD
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDA
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-253H55L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND5
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND6
 
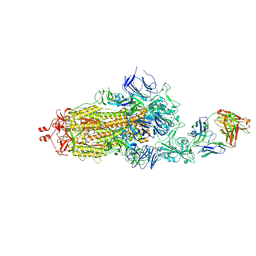 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDB
 
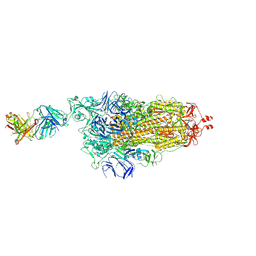 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-253H165L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
5W9C
 
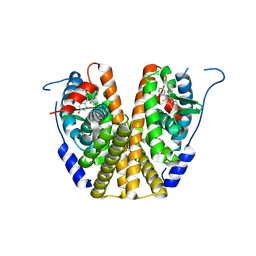 | |
2G8Q
 
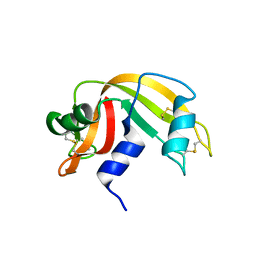 | |
3D6O
 
 | | The RNase A- 5'-Deoxy-5'-N-(ethyl isonipecotatyl)uridine complex | | Descriptor: | 1-{5-deoxy-5-[4-(ethoxycarbonyl)piperidin-1-yl]-alpha-L-arabinofuranosyl}pyrimidine-2,4(1H,3H)-dione, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-05-20 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation.
J.Med.Chem., 52, 2009
|
|
3D6Q
 
 | | The RNase A- 5'-Deoxy-5'-N-piperidinouridine complex | | Descriptor: | 1-(5-deoxy-5-piperidin-1-yl-alpha-L-arabinofuranosyl)pyrimidine-2,4(1H,3H)-dione, CITRATE ANION, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-05-20 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation.
J.Med.Chem., 52, 2009
|
|
3D6P
 
 | | RNase A- 5'-Deoxy-5'-N-morpholinouridine complex | | Descriptor: | 1-(5-deoxy-5-morpholin-4-yl-alpha-L-arabinofuranosyl)pyrimidine-2,4(1H,3H)-dione, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zogrpahos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-05-20 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation.
J.Med.Chem., 52, 2009
|
|
5TUU
 
 | |
5TUV
 
 | |
5T1Z
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Ethoxytriphenylethylene and GRIP Peptide | | Descriptor: | 4,4'-[(1Z)-1-(4-ethoxyphenyl)but-1-ene-1,2-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Rajan, S.S, Maximov, P.Y, Abderrahman, B.H, Surojeet, S, Fernandes, D.J, Fan, P, Curpan, R.F, Greene, G.L, Jordan, V.C. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Endoxifen, 4-Hydroxytamoxifen and an Estrogenic Derivative Modulate Estrogen Receptor Complex Mediated Apoptosis in Breast Cancer.
Mol. Pharmacol., 94, 2018
|
|
6MIH
 
 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
