4X5L
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 9 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AM, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9B
 
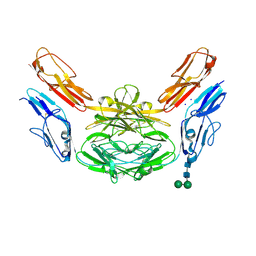 | | Crystal structure of Dscam1 isoform 4.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X83
 
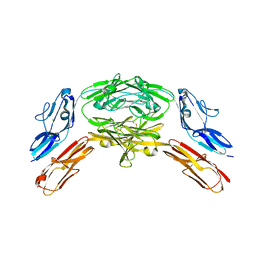 | | Crystal structure of Dscam1 isoform 7.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4WVR
 
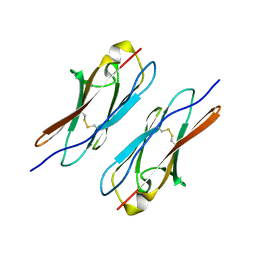 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XHQ
 
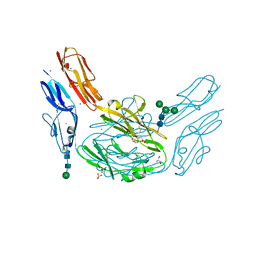 | |
4X8X
 
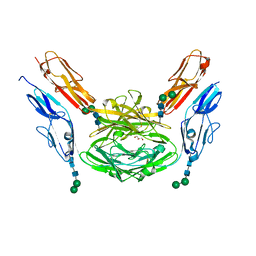 | | Crystal structure of Dscam1 isoform 1.9, N-terminal four Ig domains | | Descriptor: | Down Syndrome cell adhesion molecule isoform 1.9, GLYCEROL, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Q. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9H
 
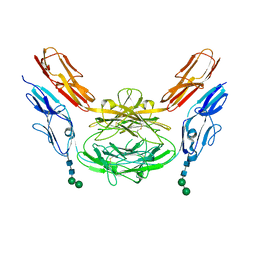 | | Crystal structure of Dscam1 isoform 8.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform AP, ... | | Authors: | Chen, Q. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
7Y54
 
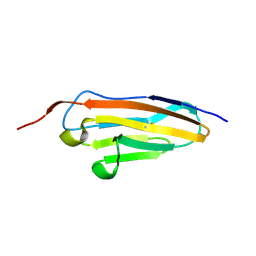 | |
7Y6O
 
 | |
7Y5J
 
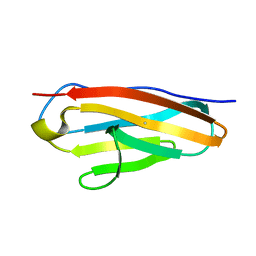 | |
7Y5R
 
 | |
7Y73
 
 | |
7Y6E
 
 | |
7Y8H
 
 | |
7Y8S
 
 | |
7Y8I
 
 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y95
 
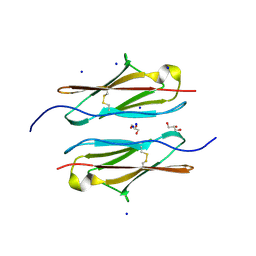 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | Descriptor: | Dscam, GLYCEROL, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y9A
 
 | | Crystal structure of sDscam Ig1-2 domains, isoform beta2v6 | | Descriptor: | Down Syndrome Cell Adhesion Molecules, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)][beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
8H2F
 
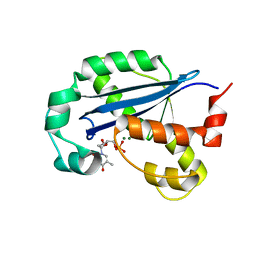 | |
8H18
 
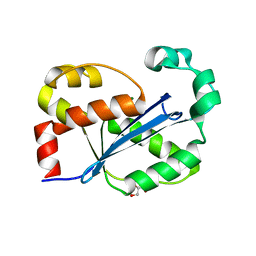 | |
4D90
 
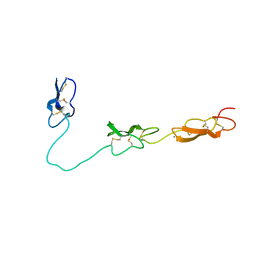 | | Crystal Structure of Del-1 EGF domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Schurpf, T, Springer, T, Wang, J. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Faseb J., 26, 2012
|
|
7Y4X
 
 | |
6JV4
 
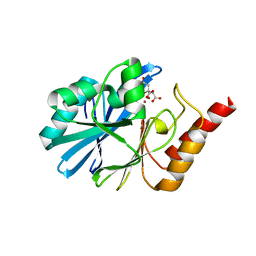 | | Crystal structure of metallo-beta-lactamase VMB-1 | | Descriptor: | CITRIC ACID, VMB-1, ZINC ION | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-04-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp.
Adv Biosyst, 4, 2020
|
|
4QH9
 
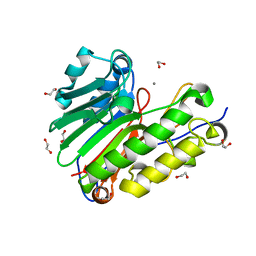 | | Crystal structure of Mn2+ bound human APE1 | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) lyase, MANGANESE (II) ION | | Authors: | Chen, Q, He, H, Georgiadis, M.M. | | Deposit date: | 2014-05-27 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I.
Biochemistry, 53, 2014
|
|
4IRE
 
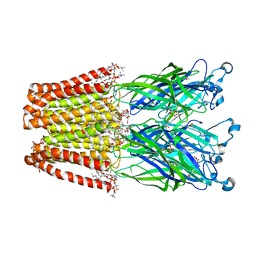 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
