4UTR
 
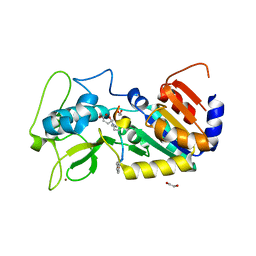 | | Crystal structure of zebrafish Sirtuin 5 in complex with glutarylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTARIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UU8
 
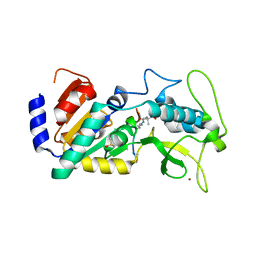 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3,3-dimethyl- succinylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 2,2-dimethylbutanedioic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UUB
 
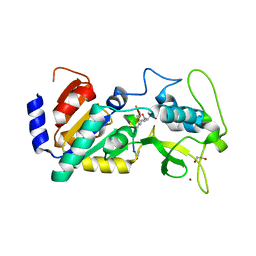 | | Crystal structure of zebrafish Sirtuin 5 in complex with 2R-butyl- succinylated CPS1-peptide | | Descriptor: | (2R)-2-butylbutanedioic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UTV
 
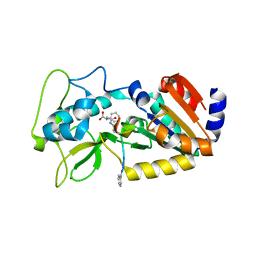 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3-phenyl- succinylated CPS1-peptide | | Descriptor: | (2R)-2-phenylbutanedioic acid, (2S)-2-phenylbutanedioic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UTX
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3-nitro- propionylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 3-NITROPROPANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UTZ
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with adipoylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBAMOYLPHOSPHATE SYNTHETASE I, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UUA
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3S-Z-amino- succinylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBAMOYLPHOSPHATE SYNTHETASE I, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5JHZ
 
 | |
2V90
 
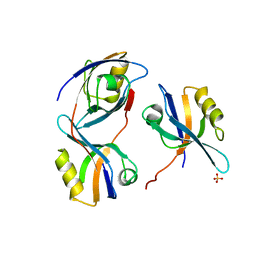 | | Crystal structure of the 3rd PDZ domain of intestine- and kidney- enriched PDZ domain IKEPP (PDZD3) | | Descriptor: | PDZ DOMAIN-CONTAINING PROTEIN 3, SULFATE ION | | Authors: | Uppenberg, J, Gileadi, C, Phillips, C, Elkins, J, Bunkoczi, G, Cooper, C, Pike, A.C.W, Salah, E, Ugochukwu, E, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D.A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the 3Rd Pdz Domain of Intestine- and Kidney-Enriched Pdz Domain Ikepp (Pdzd3)
To be Published
|
|
4UUZ
 
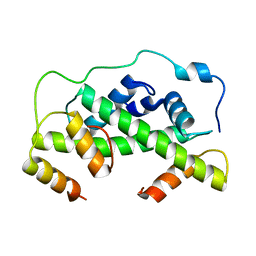 | | MCM2-histone complex | | Descriptor: | DNA REPLICATION LICENSING FACTOR MCM2, HISTONE H3, HISTONE H4 | | Authors: | Richet, N, Liu, D, Legrand, P, Bakail, M, Compper, C, Besle, A, Guerois, R, Ochsenbein, F. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Nucleic Acids Res., 43, 2015
|
|
5CZB
 
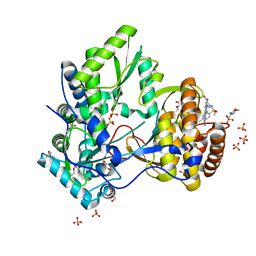 | | HCV NS5B IN COMPLEX WITH LIGAND IDX17119-5 | | Descriptor: | 1-[4-(7-amino-5-methylpyrazolo[1,5-a]pyrimidin-2-yl)phenyl]-3-{[(R)-(2,4-dimethylphenyl)(methoxy)phosphoryl]amino}-1H-pyrazole-4-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pierra, C, Dousson, C, Augustin, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Synthesis of potent and broad genotypically active NS5B HCV non-nucleoside inhibitors binding to the thumb domain allosteric site 2 of the viral polymerase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
9J8G
 
 | | mouse zbp1 hybrid complex | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*CP*A)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*G)-3'), Z-DNA-binding protein 1 | | Authors: | Gao, A.M, Zhoiu, C. | | Deposit date: | 2024-08-21 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZBP1 senses spliceosome stress through Z-RNA:DNA hybrid recognition.
Mol.Cell, 85, 2025
|
|
9JLI
 
 | | Structure of HHV6B glycoprotein B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B | | Authors: | Fang, X.Y, Xie, C, Sun, C, Zeng, M.S, Liu, Z. | | Deposit date: | 2024-09-19 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure of HHV6B glycoprotein B
To Be Published
|
|
5JUI
 
 | |
3CL3
 
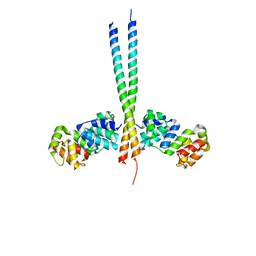 | | Crystal Structure of a vFLIP-IKKgamma complex: Insights into viral activation of the IKK signalosome | | Descriptor: | NF-kappa-B essential modulator, ORF K13 | | Authors: | Bagneris, C, Ageichik, A.V, Cronin, N, Boshoff, C, Waksman, G, Barrett, T. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome.
Mol.Cell, 30, 2008
|
|
2VNA
 
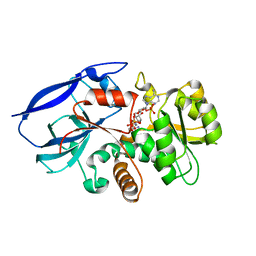 | | Structure of Human Zinc-binding Alcohol Dehydrogenase 1 (ZADH1) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROSTAGLANDIN REDUCTASE 2 | | Authors: | Shafqat, N, Kavanagh, K, Pike, A.C.W, Muniz, J.R.C, Pilka, E, Roos, A.K, Picaud, S, Johansson, C, Smee, C, Fedorov, O, Kochan, G, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, von Delft, F, Opperman, U. | | Deposit date: | 2008-02-01 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of Human Zinc-Binding Alcohol Dehydrogenase 1 (Zadh1)
To be Published
|
|
4WGI
 
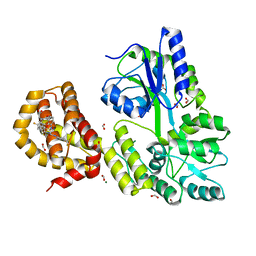 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
6F6R
 
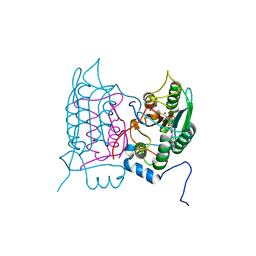 | | Crystal structure of human Caspase-1 with N-{3-[1-((S)-2-Hydroxy-5-oxo-tetrahydro-furan-3-ylcarbamoyl)-ethyl]-1-methyl-2,4-dioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl}-4-(quinoxalin-2-ylamino)-benzamide | | Descriptor: | (3~{S})-3-[[(2~{R})-2-[3-methyl-2,6-bis(oxidanylidene)-5-[[4-(quinoxalin-2-ylamino)phenyl]carbonylamino]pyrimidin-1-yl]propanoyl]amino]-4-oxidanyl-butanoic acid, Caspase-1, SULFATE ION | | Authors: | Fournier, J.F, Clary, L, Chambon, S, Dumais, L, Harris, C.S, Millois-Barbuis, C, Pierre, R, Talano, S, Thoreau, E, Aubert, J, Aurelly, M, Bouix-Peter, C, Brethon, A, Chantalat, L, Christin, O, Comino, C, El-Bazbouz, G, Ghilini, A.L, Isabet, T, Lardy, C, Luzy, A.P, Mathieu, C, Mebrouk, K, Orfila, D, Pascau, J, Reverse, K, Roche, D, Rodeschini, V, Hennequin, L.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne.
J. Med. Chem., 61, 2018
|
|
5X72
 
 | | The crystal Structure PDE delta in complex with (rac)-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, (2S)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
2IX1
 
 | | RNase II D209N mutant | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP *AP*AP*A)-3', EXORIBONUCLEASE 2, MAGNESIUM ION | | Authors: | Frazao, C, McVey, C.E, Amblar, M, Barbas, A, Vonrhein, C, Arraiano, C.M, Carrondo, M.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Unravelling the Dynamics of RNA Degradation by Ribonuclease II and its RNA-Bound Complex
Nature, 443, 2006
|
|
9LLD
 
 | | Post-fusion ectodomain of KSHV gB in complex with 2C4 Fab | | Descriptor: | 2C4 Fab H chain, 2C4 Fab L chain, Core gene UL27 family protein | | Authors: | Fang, X.Y, Sun, C, Xie, C, Zeng, M.S, Liu, Z. | | Deposit date: | 2025-01-17 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Post-fusion ectodomain of KSHV gB in complex with 2C4 Fab
To Be Published
|
|
3D21
 
 | | Crystal structure of a poplar wild-type thioredoxin h, PtTrxh4 | | Descriptor: | Thioredoxin H-type | | Authors: | Koh, C.S, Didierjean, C, Corbier, C, Rouhier, N, Jacquot, J.P, Gelhaye, E. | | Deposit date: | 2008-05-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Atypical Catalytic Mechanism Involving Three Cysteines of Thioredoxin.
J.Biol.Chem., 283, 2008
|
|
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
