4IM8
 
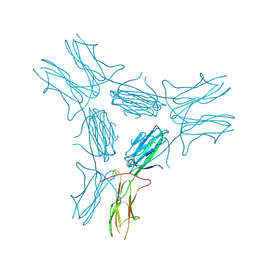 | | low resolution crystal structure of mouse RAGE | | Descriptor: | Advanced glycation end-products receptor | | Authors: | Xu, D, Young, J.H, Krahn, J.M, Song, D, Corbett, K.D, Chazin, W.J, Pedersen, L.C, Esko, J.D. | | Deposit date: | 2013-01-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction.
Acs Chem.Biol., 8, 2013
|
|
4IPC
 
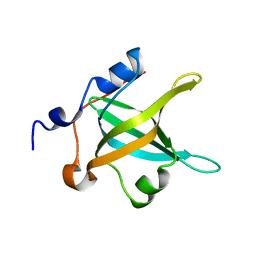 | | Structure of the N-terminal domain of RPA70, E7R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
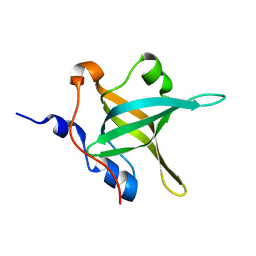 | | Structure of the N-terminal domain of RPA70, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPG
 
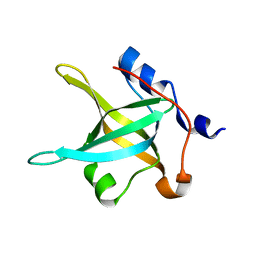 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
1PSB
 
 | | Solution structure of calcium loaded S100B complexed to a peptide from N-Terminal regulatory domain of NDR kinase. | | Descriptor: | Ndr Ser/Thr kinase-like protein, S-100 protein, beta chain | | Authors: | Bhattacharya, S, Large, E, Heizmann, C.W, Hemmings, B, Chazin, W.J. | | Deposit date: | 2003-06-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Biochemistry, 42, 2003
|
|
1QMS
 
 | | Head-to-Tail Dimer of Calicheamicin gamma-1-I Oligosaccharide Bound to DNA Duplex, NMR, 9 Structures | | Descriptor: | CALICHEAMICIN GAMMA-1-OLIGOSACCHARIDE, DNA (5'-D(*GP*CP*AP*CP*CP*TP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*AP*GP*GP*TP*GP*C)-3'), ... | | Authors: | Bifulco, G, Galeone, A, Nicolaou, K.C, Chazin, W.J, Gomez-Paloma, L. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Complex between the Head-to-Tail Dimer of Calicheamicin Gamma-1-I Oligosaccharide and a DNA Duplex Containing D(ACCT) and D(TCCT) High-Affinity Binding Sites
J.Am.Chem.Soc., 120, 1998
|
|
1QX2
 
 | | X-ray Structure of Calcium-loaded Calbindomodulin (A Calbindin D9k Re-engineered to Undergo a Conformational Opening) at 1.44 A Resolution | | Descriptor: | CALCIUM ION, Vitamin D-dependent calcium-binding protein, intestinal, ... | | Authors: | Bunick, C.G, Nelson, M.R, Mangahas, S, Mizoue, L.S, Bunick, G.J, Chazin, W.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-05-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Designing Sequence to Control Protein Function in an EF-Hand Protein
J.Am.Chem.Soc., 126, 2004
|
|
1OQP
 
 | |
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
4LIL
 
 | | Crystal structure of the catalytic subunit of human primase bound to UTP and Mn | | Descriptor: | DNA primase small subunit, MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Vaithiyalingam, S, Eichman, B.F, Chazin, W.J. | | Deposit date: | 2013-07-02 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Eukaryotic Primer Synthesis from Structures of the p48 Subunit of Human DNA Primase.
J.Mol.Biol., 426, 2014
|
|
4LIK
 
 | |
4LIM
 
 | | Crystal structure of the catalytic subunit of yeast primase | | Descriptor: | DNA primase small subunit, ZINC ION | | Authors: | Vaithiyalingam, S, Chazin, W.J, Berger, J.M, Corn, J, Stephenson, S. | | Deposit date: | 2013-07-02 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into Eukaryotic Primer Synthesis from Structures of the p48 Subunit of Human DNA Primase.
J.Mol.Biol., 426, 2014
|
|
4LUV
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 1-(3-methylphenyl)-5-phenyl-1H-pyrazole-3-carboxylic acid, 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LW1
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LWC
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-[3-chloro-4-({4-[1-(3,4-dichlorophenyl)-1H-pyrazol-5-yl]benzyl}carbamothioyl)phenyl]furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LUZ
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LUO
 
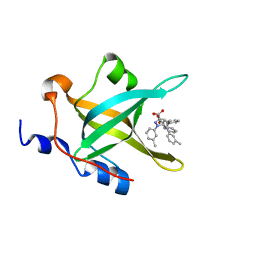 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 1-(3-methylphenyl)-5-phenyl-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
1CNP
 
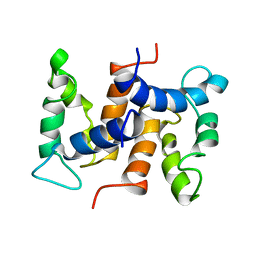 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
1CB1
 
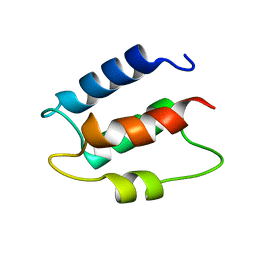 | |
1DSI
 
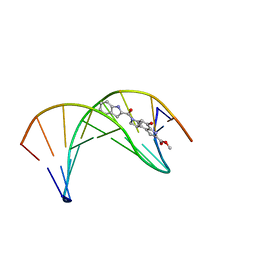 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
1DPU
 
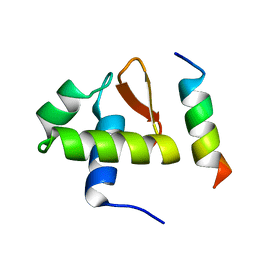 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN RPA32 COMPLEXED WITH UNG2(73-88) | | Descriptor: | REPLICATION PROTEIN A (RPA32) C-TERMINAL DOMAIN, URACIL DNA GLYCOSYLASE (UNG2) | | Authors: | Mer, G, Edwards, A.M, Chazin, W.J. | | Deposit date: | 1999-12-27 | | Release date: | 2000-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA.
Cell(Cambridge,Mass.), 103, 2000
|
|
1DSM
 
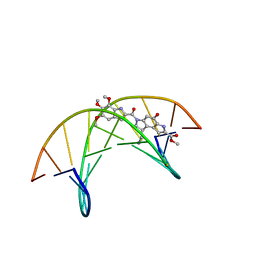 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
1DSA
 
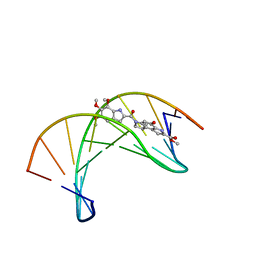 | | (+)-DUOCARMYCIN SA COVALENTLY LINKED TO DUPLEX DNA, NMR, 20 STRUCTURES | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Eis, P.S, Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA.
J.Mol.Biol., 272, 1997
|
|
1CQO
 
 | |
