1CZF
 
 | | ENDO-POLYGALACTURONASE II FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, POLYGALACTURONASE II, ZINC ION | | Authors: | van Santen, Y, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-02 | | Release date: | 1999-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis.
J.Biol.Chem., 274, 1999
|
|
1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
167L
 
 | |
180L
 
 | |
175L
 
 | |
1A16
 
 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AI7
 
 | | PENICILLIN ACYLASE COMPLEXED WITH PHENOL | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, PHENOL | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
169L
 
 | |
8BV3
 
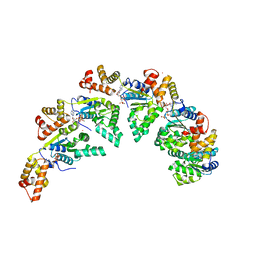 | | Bacillus subtilis DnaA domain III structure | | Descriptor: | Chromosomal replication initiator protein DnaA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pintar, S, Hubbard, J.A. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The bacterial replication origin BUS promotes nucleobase capture.
Nat Commun, 14, 2023
|
|
168L
 
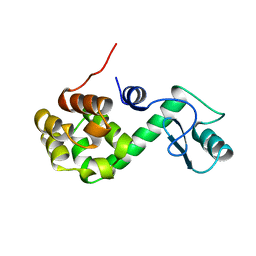 | |
1A00
 
 | | HEMOGLOBIN (VAL BETA1 MET, TRP BETA37 TYR) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1A0Z
 
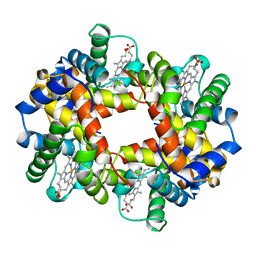 | | HEMOGLOBIN (VAL BETA1 MET) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1AK5
 
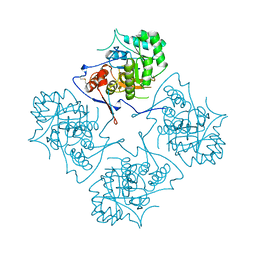 | |
178L
 
 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
1YOU
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with a potent pyrimidinetrione inhibitor | | Descriptor: | 5-(2-ETHOXYETHYL)-5-[4-(4-FLUOROPHENOXY)PHENOXY]PYRIMIDINE-2,4,6(1H,3H,5H)-TRIONE, CALCIUM ION, Collagenase 3, ... | | Authors: | Pandit, J. | | Deposit date: | 2005-01-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent pyrimidinetrione-based inhibitors of MMP-13 with enhanced selectivity over MMP-14.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1BU7
 
 | | CRYOGENIC STRUCTURE OF CYTOCHROME P450BM-3 HEME DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYTOCHROME P450), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BN3
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFINAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNN
 
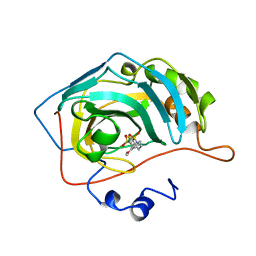 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 3,,4-DIHYDRO-2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNV
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (S)-3,4-DIHYDRO-2-(3-METHOXYPHENYL)-4-METHYLAMINO-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1D2U
 
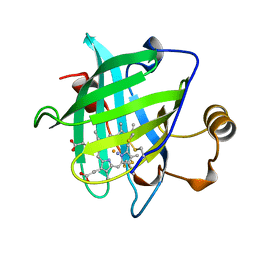 | | 1.15 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 1999-09-28 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1CPY
 
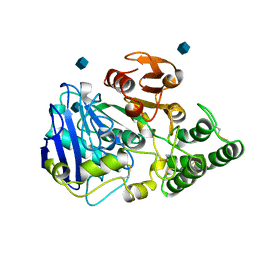 | | SITE-DIRECTED MUTAGENESIS ON (SERINE) CARBOXYPEPTIDASE Y FROM YEAST. THE SIGNIFICANCE OF THR 60 AND MET 398 IN HYDROLYSIS AND AMINOLYSIS REACTIONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE | | Authors: | Sorensen, S.B, Raaschou-Nielsen, M, Mortensen, U, Remington, S.J, Breddam, K. | | Deposit date: | 1995-03-24 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Site-Directed Mutagenesis on (Serine) Carboxypeptidase Y from Yeast. The Significance of Thr 60 and met 398 in Hydrolysis and Aminolysis Reactions
J.Am.Chem.Soc., 117, 1995
|
|
1XFG
 
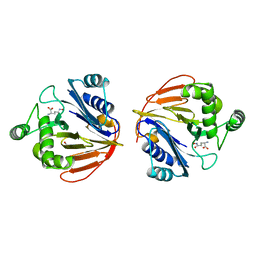 | | Glutaminase domain of glucosamine 6-phosphate synthase complexed with l-glu hydroxamate | | Descriptor: | ACETATE ION, GLUTAMINE HYDROXAMATE, Glucosamine--fructose-6-phosphate aminotransferase [isomerizing], ... | | Authors: | Isupov, M.N, Teplyakov, A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Binding is Required for Assembly of the Active Conformation of the Catalytic Site in Ntn Amidotransferases: Evidence from the 1.8 Angstrom Crystal Structure of the Glutaminase Domain of Glucosamine 6-Phosphate Synthase
Structure, 4, 1996
|
|
1D0O
 
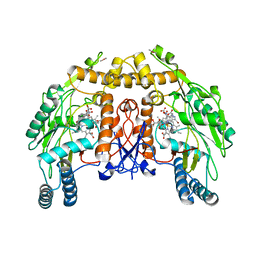 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 3-BROMO-7-NITROINDAZOLE (H4B PRESENT) | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, ACETATE ION, BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-13 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
1AJQ
 
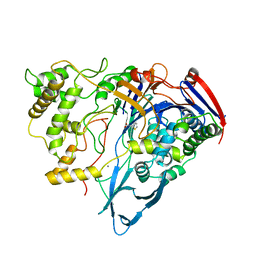 | | PENICILLIN ACYLASE COMPLEXED WITH THIOPHENEACETIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, THIOPHENEACETIC ACID | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
176L
 
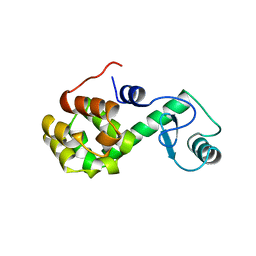 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L, Dubose, R, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
