5JY3
 
 | |
4ZFC
 
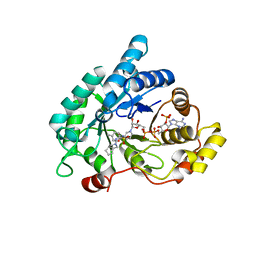 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
5ZWZ
 
 | |
5ZWX
 
 | |
6W78
 
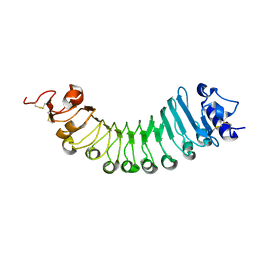 | | crystal structure of a plant ice-binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antifreeze polypeptide | | Authors: | Wang, Y.N, Zhang, H.Q. | | Deposit date: | 2020-03-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Carrot 'antifreeze' protein has an irregular ice-binding site that confers weak freezing point depression but strong inhibition of ice recrystallization.
Biochem.J., 477, 2020
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
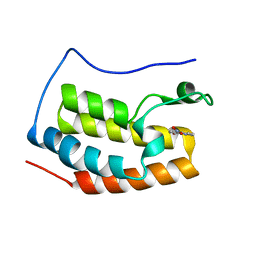
5Y1Y
 
 | |
7FAT
 
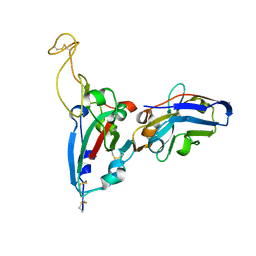 | | Structure Determination of the RBD-NB1A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | Authors: | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
8I3E
 
 | |
6Z01
 
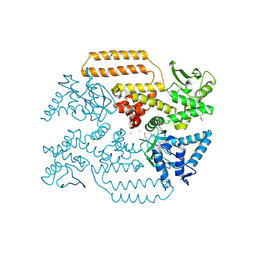 | | DNA Topoisomerase | | Descriptor: | CHLORIDE ION, DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
6Z03
 
 | | DNA Topoisomerase | | Descriptor: | DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
1N9A
 
 | | Farnesyltransferase complex with tetrahydropyridine inhibitors | | Descriptor: | 1-{2-[3-(4-CYANO-BENZYL)-3H-IMIDAZOL-4-YL]-ACETYL}-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: bioavailable analogues with improved cellular potency.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4RC8
 
 | |
1C9Q
 
 | |
1N95
 
 | | Aryl Tetrahydrophyridine Inhbitors of Farnesyltranferase: Glycine, Phenylalanine and Histidine Derivatives | | Descriptor: | 1-[2-(4-CYANO-BENZYLAMINO)-3-(3-METHYL-3H-IMIDAZOL-4-YL)-PROPIONYL]-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4RC5
 
 | |
4RI2
 
 | | Crystal structure of the photoprotective protein PsbS from spinach | | Descriptor: | CHLOROPHYLL A, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RI3
 
 | | Crystal structure of DCCD-modified PsbS from spinach | | Descriptor: | DICYCLOHEXYLUREA, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1N94
 
 | | Aryl Tetrahydropyridine Inhbitors of Farnesyltransferase: Glycine, Phenylalanine and Histidine Derivates | | Descriptor: | 2-{(5-{[BUTYL-(2-CYCLOHEXYL-ETHYL)-AMINO]-METHYL}-2'-METHYL-BIPHENYL-2-CARBONYL)-AMINO]-4-METHYLSULFANYL-BUTYRIC ACID, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Connor, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4QUW
 
 | |
4RC7
 
 | |
4YPQ
 
 | | Crystal structure of the ROR(gamma)t ligand binding domain in complex with 4-(1-(2-chloro-6-(trifluoromethyl)benzoyl)-1H-indazol-3-yl)benzoic acid | | Descriptor: | 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, MAGNESIUM ION, Nuclear receptor ROR-gamma | | Authors: | Leysen, S, Scheepstra, M, van Almen, G.C, Ottmann, C, Brunsveld, L. | | Deposit date: | 2015-03-13 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of an allosteric binding site for ROR gamma t inhibition.
Nat Commun, 6, 2015
|
|
3OCG
 
 | | P38 Alpha kinase complexed with a 5-amino-pyrazole based inhibitor | | Descriptor: | 5-amino-N-[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]-1-phenyl-1H-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 5-Amino-pyrazoles as potent and selective p38α inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
9B7A
 
 | | Crystal structure of peroxiredoxin 1 with RA | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, Peroxiredoxin-1 | | Authors: | Wu, Y, Xu, H, Luo, C. | | Deposit date: | 2024-03-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inhibited peroxidase activity of peroxiredoxin 1 by palmitic acid exacerbates nonalcoholic steatohepatitis in male mice.
Nat Commun, 16, 2025
|
|
5S9R
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMS-986158, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
