8DM0
 
 | | Cryo-EM structure of SARS-CoV-2 D614G spike protein in complex with VH ab6 (focused refinement of NTD and VH ab6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab6 | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLI
 
 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLL
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLQ
 
 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLZ
 
 | | Cryo-EM structure of SARS-CoV-2 D614G spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
6JEG
 
 | |
8JXZ
 
 | | Chitin binding SusD-like protein AqSusD in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SusD-like protein AqSusD | | Authors: | Yang, J. | | Deposit date: | 2023-07-01 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
6JCO
 
 | |
7SSK
 
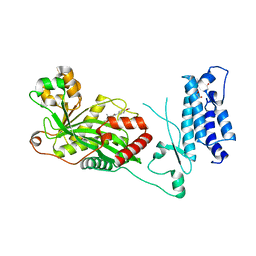 | | Human P300 complexed with a glycine-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SZQ
 
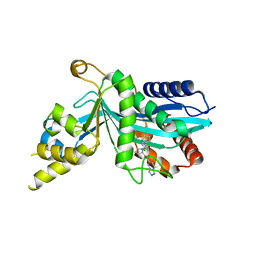 | | Human P300 complexed with an azaindazole inhibitor | | Descriptor: | 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-1H-pyrazolo[4,3-b]pyridin-5-yl-D-prolinamide, Histone acetyltransferase p300 | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SS8
 
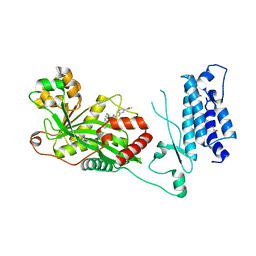 | | Human P300 complexed with a proline-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-{[3-(methylcarbamoyl)phenyl]methyl}-D-prolinamide, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
8T62
 
 | |
8T61
 
 | |
8T63
 
 | |
3GHW
 
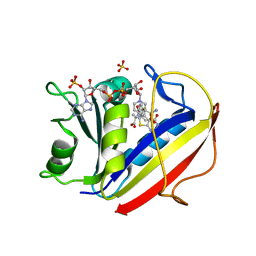 | | Human dihydrofolate reductase inhibitor complex | | Descriptor: | Dihydrofolate reductase, N-({4-[(2-amino-6-methyl-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-5-yl)sulfanyl]phenyl}carbonyl)-L-glutamic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2009-03-04 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Design, synthesis, and X-ray crystal structure of classical and nonclassical 2-amino-4-oxo-5-substituted-6-ethylthieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors and as potential antitumor agents
J.Med.Chem., 52, 2009
|
|
3GI2
 
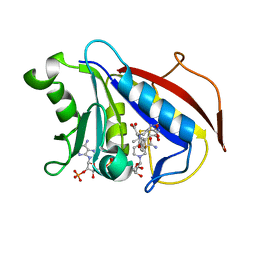 | | Human dihydrofolate reductase Q35K mutant inhibitor complex | | Descriptor: | Dihydrofolate reductase, N-({4-[(2-amino-6-methyl-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-5-yl)sulfanyl]phenyl}carbonyl)-L-glutamic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design, synthesis, and X-ray crystal structure of classical and nonclassical 2-amino-4-oxo-5-substituted-6-ethylthieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors and as potential antitumor agents.
J.Med.Chem., 52, 2009
|
|
1Q4Q
 
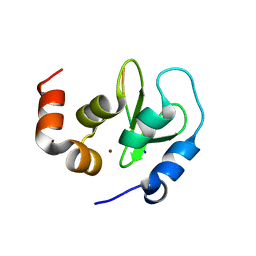 | | Crystal structure of a DIAP1-Dronc complex | | Descriptor: | Apoptosis 1 inhibitor, Nedd2-like caspase CG8091-PA, ZINC ION | | Authors: | Chai, J, Yan, N, Shi, Y. | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination
Nat.Struct.Biol., 10, 2003
|
|
3GHV
 
 | | Human dihydrofolate reductase Q35K/N64F double mutant inhibitor complex | | Descriptor: | Dihydrofolate reductase, GLYCEROL, N-({4-[(2-amino-6-ethyl-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-5-yl)sulfanyl]phenyl}carbonyl)-L-glutamic acid, ... | | Authors: | Cody, V. | | Deposit date: | 2009-03-04 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design, synthesis, and X-ray crystal structure of classical and nonclassical 2-amino-4-oxo-5-substituted-6-ethylthieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors and as potential antitumor agents
J.Med.Chem., 52, 2009
|
|
8YKQ
 
 | |
8YKP
 
 | |
8YKN
 
 | |
8YKM
 
 | |
8YKL
 
 | | Crystal structure of MERS main protease in complex with X77 | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77.
Int.J.Biol.Macromol., 276, 2024
|
|
8YKK
 
 | | Crystal structure of SARS main protease in complex with X77 | | Descriptor: | 3C-like proteinase nsp5, N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77.
Int.J.Biol.Macromol., 276, 2024
|
|
1SDZ
 
 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
