1BEQ
 
 | |
1BES
 
 | |
1BEJ
 
 | |
1BEM
 
 | |
8AM3
 
 | |
8AM8
 
 | |
8AM6
 
 | |
8GP6
 
 | |
2FT6
 
 | |
2FT8
 
 | |
2FT7
 
 | |
2FTA
 
 | |
1D41
 
 | |
7ZWQ
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 10 | | Descriptor: | 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWY
 
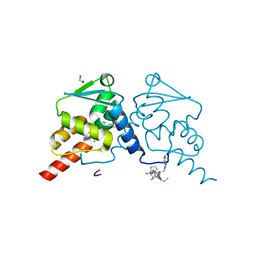 | | Crystal structure of human BCL6 BTB domain in complex with compound 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(phenylmethyl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Shetty, K, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWN
 
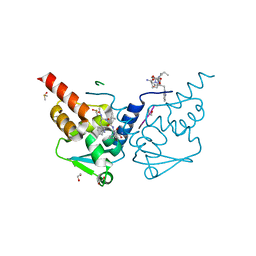 | | Crystal structure of human BCL6 BTB domain in complex with a WVIP peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Shetty, K, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWW
 
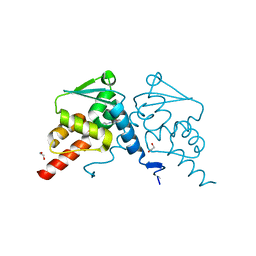 | | Crystal structure of human BCL6 BTB domain in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-5-[(6-morpholin-4-ylpyrimidin-4-yl)amino]benzimidazol-2-one, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWP
 
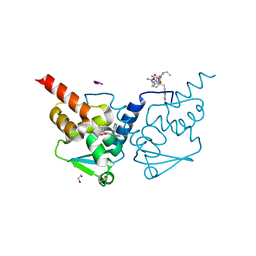 | | Crystal structure of human BCL6 BTB domain in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Shetty, K, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWT
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-chlorophenyl)amino]-~{N}-(pyridin-2-ylmethyl)-1,3-thiazole-4-carboxamide, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7B9L
 
 | | MEK1 in complex with compound 23 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B94
 
 | | MEK1 in complex with compound 6 | | Descriptor: | 2-(4-iodophenyl)-8~{H}-imidazo[1,2-c]pyrimidin-5-one, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7C20
 
 | |
7C21
 
 | | Crystal structure of Duvenhage virus phosphoprotein C-terminal domain | | Descriptor: | Phosphoprotein | | Authors: | Sugiyama, A, Jiang, X, Maenaka, K, Yao, M, Ose, T. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of the C-terminal domain of functionally divergent lyssavirus P proteins.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7C7P
 
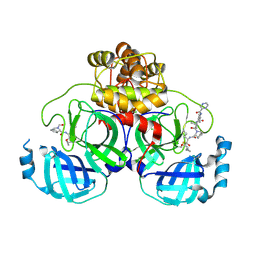 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
