7CIB
 
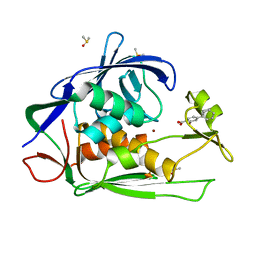 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-oxidanyl-4-phenyl-benzoic acid, DIMETHYL SULFOXIDE, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7B7V
 
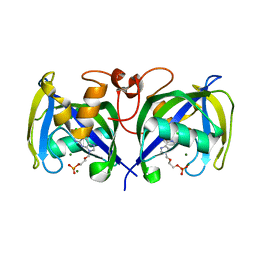 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
6LBH
 
 | |
3WZY
 
 | |
3WZX
 
 | |
3WZW
 
 | |
3WZV
 
 | |
2E5T
 
 | |
2E5U
 
 | |
2E5Y
 
 | |
2ZY6
 
 | | Crystal structure of a truncated tRNA, TPHE39A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Tanaka, I, Yao, M, Tanaka, Y, Kitago, Y, Ymagata, S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand
Rna, 15, 2009
|
|
3DSD
 
 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3DSC
 
 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
1DYK
 
 | | Laminin alpha 2 chain LG4-5 domain pair | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Tisi, D, Talts, J.F, Timple, R, Hohenester, E. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-Terminal Laminin G-Like Domain Pair of the Laminin Alpha 2 Chain Harbouring Binding Sites for Alpha-Dystroglycan and Heparin
Embo J., 19, 2000
|
|
5GU5
 
 | |
2A71
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), orthorhombic crystal form | | Descriptor: | Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Y
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), tetragonal crystal form | | Descriptor: | Emp47p (form1), SULFATE ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6V
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), potassium-bound form | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Z
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 1 | | Descriptor: | Emp47p (form2) | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A70
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6W
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), metal-free form | | Descriptor: | Emp46p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6X
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), Y131F mutant | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
1OKQ
 
 | | LAMININ ALPHA 2 CHAIN LG4-5 DOMAIN PAIR, CA1 SITE MUTANT | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Wizemann, H, Garbe, J.H.O, Friedrich, M.V.K, Timpl, R, Sasaki, T, Hohenester, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distinct Requirements for Heparin and Alpha-Dystroglycan Binding Revealed by Structure-Based Mutagenesis of the Laminin Alpha2 Lg4-Lg5 Domain Pair
J.Mol.Biol., 332, 2003
|
|
1ERZ
 
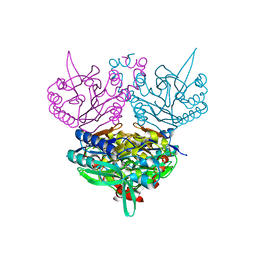 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | Descriptor: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | Authors: | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | Deposit date: | 2000-04-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
3HUF
 
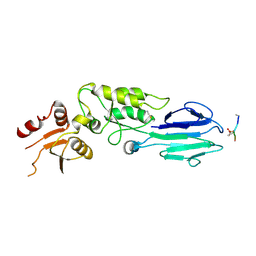 | | Structure of the S. pombe Nbs1-Ctp1 complex | | Descriptor: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | Authors: | Williams, R.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-06-13 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
