1CYZ
 
 | | NMR STRUCTURE OF THE GAACTGGTTC/TRI-IMIDAZOLE POLYAMIDE COMPLEX | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, 5'-D(*GP*AP*AP*CP*TP*GP*GP*TP*TP*C)-3' | | Authors: | Yang, X.-L, Hubbard IV, R.B, Lee, M, Tao, Z.-F, Sugiyama, H, Wang, A.H.-J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Imidazole-imidazole pair as a minor groove recognition motif for T:G mismatched base pairs
Nucleic Acids Res., 27, 1999
|
|
1D38
 
 | |
4BE6
 
 | |
1UAQ
 
 | | The crystal structure of yeast cytosine deaminase | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, ZINC ION, cytosine deaminase | | Authors: | Ko, T.-P, Lin, J.-J, Hu, C.-Y, Hsu, Y.-H, Wang, A.H.-J, Liaw, S.-H. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution
J.Biol.Chem., 278, 2003
|
|
1V7U
 
 | | Crystal structure of Undecaprenyl Pyrophosphate Synthase with farnesyl pyrophosphate | | Descriptor: | FARNESYL DIPHOSPHATE, Undecaprenyl pyrophosphate synthetase | | Authors: | Chang, S.-Y, Ko, T.-P, Chen, A.P.-C, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2003-12-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies
Protein Sci., 13, 2004
|
|
1VG2
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG6
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A/I123A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VQB
 
 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Skinner, M.M, Zhang, H, Leschnitzer, D.H, Guan, Y, Bellamy, H, Sweet, R.M, Gray, C.W, Konings, R.N.H, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the gene V protein of bacteriophage f1 determined by multiwavelength x-ray diffraction on the selenomethionyl protein.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1YHB
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1YHA
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
3AHY
 
 | | Crystal structure of beta-glucosidase 2 from fungus Trichoderma reesei in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
3AHZ
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, GLYCEROL | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
3AHX
 
 | |
3AI0
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with para-nitrophenyl-beta-D-glucopyranoside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, GLYCEROL, beta-glucosidase | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
1DSD
 
 | |
1DSC
 
 | |
2DTN
 
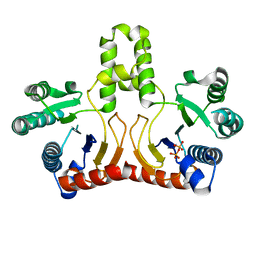 | | Crystal structure of Helicobacter pylori undecaprenyl pyrophosphate synthase complexed with pyrophosphate | | Descriptor: | DIPHOSPHATE, undecaprenyl pyrophosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chen, C.L, Ko, T.P, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical characterization, crystal structure, and inhibitors of Helicobacter pylori undecaprenyl pyrophosphate synthase
To be Published
|
|
2E11
 
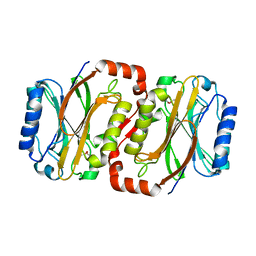 | | The Crystal Structure of XC1258 from Xanthomonas campestris: A CN-hydrolase Superfamily Protein with an Arsenic Adduct in the Active Site | | Descriptor: | CACODYLATE ION, Hydrolase | | Authors: | Chin, K.-H, Tsai, Y.-D, Chan, N.-L, Huang, K.-F, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of XC1258 from Xanthomonas campestris: A putative procaryotic Nit protein with an arsenic adduct in the active site
Proteins, 69, 2007
|
|
2E94
 
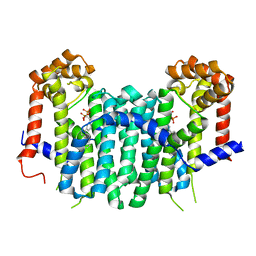 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-364 | | Descriptor: | 3-BIPHENYL-3-YL-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3RNZ
 
 | |
2E95
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-675 | | Descriptor: | 1-HYDROXY-2-[3'-(NAPHTHALENE-2-SULFONYLAMINO)-BIPHENYL-3-YL]ETHYLIDENE-1,1-BISPHOSPHONIC ACID, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Cao, R, Ko, T.P, Jeng, W.Y, Chang, T.H, Chen, C.K.-M, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3RO0
 
 | |
2E8V
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with product GGPP (P21) | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase | | Authors: | Chen, C.K.-M, Guo, R.T, Ko, T.P, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E92
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-261 | | Descriptor: | (1-HYDROXY-2-IMIDAZO[1,2-A]PYRIDIN-3-YLETHANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Jeng, W.Y, Chen, C.K.-M, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E8U
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and IPP (P21) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Chen, C.K.-M, Ko, T.P, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
