5B36
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
3QFZ
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiose Phosphorylase, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3QFY
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Cellobiose Phosphorylase, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3QG0
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Cellobiose Phosphorylase, PHOSPHATE ION, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
1D8L
 
 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
6AGZ
 
 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
4ZOB
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZOE
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZO9
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZOA
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, DI(HYDROXYETHYL)ETHER, Lin1840 protein, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZO8
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorose | | Descriptor: | Lin1840 protein, MAGNESIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZO6
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with cellobiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZO7
 
 | | Crystal structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with gentiobiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZOC
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorotriose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZOD
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lin1840 protein, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4H8N
 
 | | Crystal structure of conjugated polyketone reductase C2 from candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3AK4
 
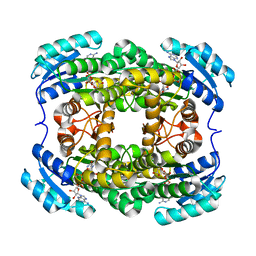 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
2ZAM
 
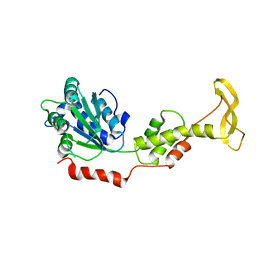 | | Crystal structure of mouse SKD1/VPS4B apo-form | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZAO
 
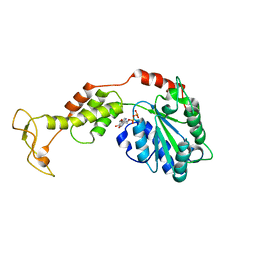 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
4KTP
 
 | | Crystal structure of 2-O-alpha-glucosylglycerol phosphorylase in complex with glucose | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 65 central catalytic, PENTAETHYLENE GLYCOL, ... | | Authors: | Touhara, K.K, Nihira, T, Kitaoka, M, Nakai, H, Fushinobu, S. | | Deposit date: | 2013-05-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-alpha-glucosylglycerol phosphorylase
J.Biol.Chem., 289, 2014
|
|
4KTR
 
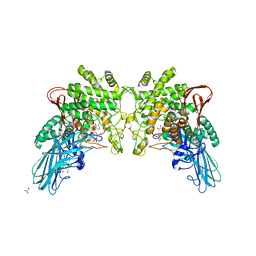 | | Crystal structure of 2-O-alpha-glucosylglycerol phosphorylase in complex with isofagomine and glycerol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Touhara, K.K, Nihira, T, Kitaoka, M, Nakai, H, Fushinobu, S. | | Deposit date: | 2013-05-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-alpha-glucosylglycerol phosphorylase
J.Biol.Chem., 289, 2014
|
|
2ZAN
 
 | | Crystal structure of mouse SKD1/VPS4B ATP-form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
1V4R
 
 | | Solution structure of Streptmycal repressor TraR | | Descriptor: | Transcriptional Repressor | | Authors: | Tanaka, T, Komatsu, C, Kobayashi, K, Sugai, M, Kataoka, M, Kohno, T. | | Deposit date: | 2003-11-17 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Streptmycal repressor TraR
TO BE PUBLISHED
|
|
3WGB
 
 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2D02
 
 | | R52Q Mutant of Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
