6YXU
 
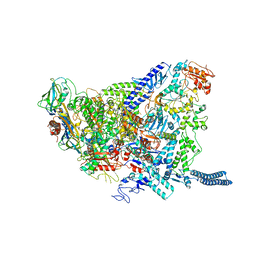 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State I, primary channel engaged | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
6YYS
 
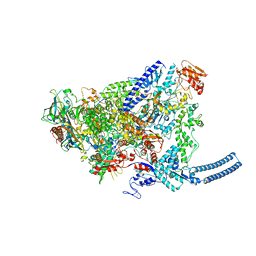 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State II, primary channel engaged and active site interfering | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
3M9Z
 
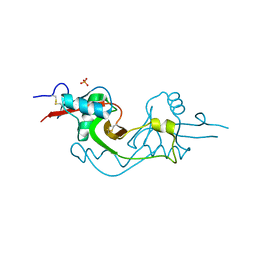 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
1M0B
 
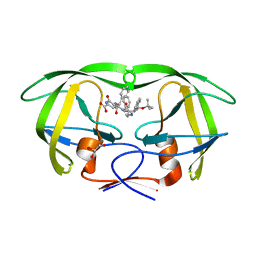 | | HIV-1 protease in complex with an ethyleneamine inhibitor | | Descriptor: | GLYCEROL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Petrokova, H, Hasek, J, Dohnalek, J. | | Deposit date: | 2002-06-12 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of hydroxyl group and R/S configuration of isostere in binding properties of HIV-1 protease inhibitors
Eur.J.Biochem., 271, 2004
|
|
8QJL
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia | | Descriptor: | GLYCEROL, S1/P1 Nuclease, SULFATE ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJN
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with adenosine-5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJO
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with guanosine-5'-monophosphate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJP
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with uridine - 5'- monophosphate | | Descriptor: | GLYCEROL, PHOSPHATE ION, S1/P1 Nuclease, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJM
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, PENTAETHYLENE GLYCOL, S1/P1 Nuclease, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJQ
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with cytidine - 5' - monophosphate as an inhibitor. | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
5FBB
 
 | | S1 nuclease from Aspergillus oryzae in complex with phosphate and adenosine 5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5FB9
 
 | | S1 nuclease from Aspergillus oryzae with unoccupied active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5FBC
 
 | | S1 nuclease from Aspergillus oryzae in complex with 2'-deoxyadenosine-5'-thio-monophosphate (5'dAMP(S)). | | Descriptor: | 2-DEOXY-ADENOSINE -5'-THIO-MONOPHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5FBF
 
 | | S1 nuclease from Aspergillus oryzae in complex with two molecules of 2'-deoxycytidine-5'-monophosphate | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5FBA
 
 | | S1 nuclease from Aspergillus oryzae in complex with phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5FBD
 
 | | S1 nuclease from Aspergillus oryzae in complex with phosphate and 2'-deoxycytidine | | Descriptor: | 2'-DEOXYCYTIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
