6SXR
 
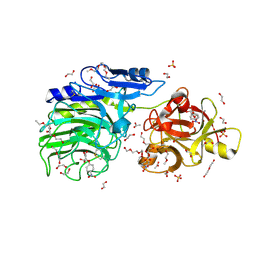 | | E221Q mutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
6SXS
 
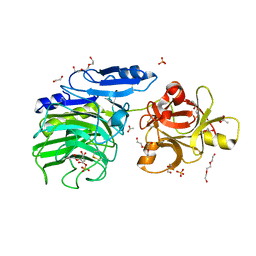 | | GH54 a-l-arabinofuranosidase soaked with cyclic sulfate inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
6SXT
 
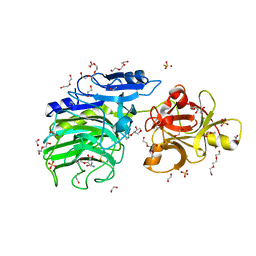 | | GH54 a-l-arabinofuranosidase soaked with aziridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
5M7I
 
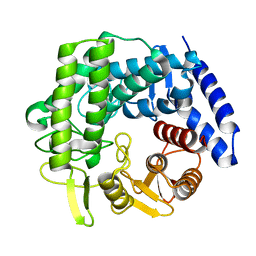 | | Crystal structure of GH125 1,6-alpha-mannosidase mutant from Clostridium perfringens in complex with 1,6-alpha-mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, exo-alpha-1,6-mannosidase | | Authors: | Males, A, Alonso-Gil, S, Fernandes, P, Williams, S.J, Rovira, C, Davies, G.J. | | Deposit date: | 2016-10-27 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational Design of Experiment Unveils the Conformational Reaction Coordinate of GH125 alpha-Mannosidases.
J. Am. Chem. Soc., 139, 2017
|
|
5NDX
 
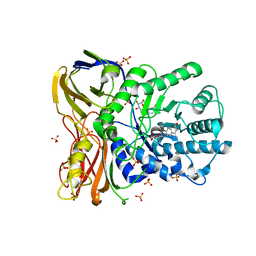 | | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyl hydrolase, SULFATE ION | | Authors: | Raich, L, Valero-Gonzalez, J, Castro-Lopez, J, Millan, C, Jimenez-Garcia, M.J, Nieto, P, Uson, I, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2017-03-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent:
Crystallography and QM/MM insights into Mucopolysaccharidosis I.
To Be Published
|
|
5M03
 
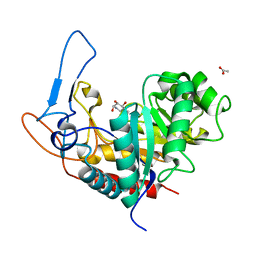 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin and 1,2-alpha-mannobiose | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5M3W
 
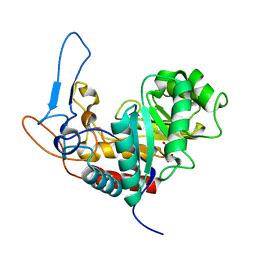 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5M17
 
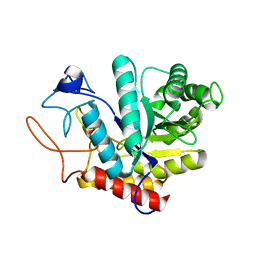 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5MC8
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5LYR
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5M5D
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5MEL
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with Glc-alpha-1,3-(3R,4R,5R)-5-(hydroxymethyl)cyclohex-1,2-ene-3,4-diol | | Descriptor: | (1~{R},2~{R},6~{R})-6-(hydroxymethyl)cyclohex-3-ene-1,2-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5M7Y
 
 | | Crystal structure of GH125 1,6-alpha-mannosidase mutant from Clostridium perfringens in complex with 1,6-alpha-mannotriose | | Descriptor: | 1,6-alpha-mannosidase, MAGNESIUM ION, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose | | Authors: | Males, A, Alonso-Gil, S, Fernandes, P, Williams, S.J, Rovira, C, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Computational Design of Experiment Unveils the Conformational Reaction Coordinate of GH125 alpha-Mannosidases.
J. Am. Chem. Soc., 139, 2017
|
|
6EQJ
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant, apo form | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1 | | Authors: | Bailey, H.J, Kopec, J, Bilyard, M.K, Bezerra, G.A, Seo Lee, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Davis, B.G, Yue, W.W. | | Deposit date: | 2017-10-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis.
Nature, 563, 2018
|
|
5JTS
 
 | | Structure of a beta-1,4-mannanase, SsGH134. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
3F9P
 
 | | Crystal structure of myeloperoxidase from human leukocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Carpena, X, Fita, I, Obinger, C. | | Deposit date: | 2008-11-14 | | Release date: | 2009-07-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Essential role of proximal histidine-asparagine interaction in Mammalian peroxidases.
J.Biol.Chem., 284, 2009
|
|
8RBI
 
 | |
9EOB
 
 | | Engineered GH181 sialidase from Akkermansia muciniphila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pichler, M.J, Banerjee, S, Nielsen, T.S, Morth, J.P, Abou Hachem, M. | | Deposit date: | 2024-03-14 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineered GH181 sialidase from Akkermansia muciniphila
Acs Catalysis, 2025
|
|
3D6E
 
 | | Crystal structure of the engineered 1,3-1,4-beta-glucanase protein from Bacillus licheniformis | | Descriptor: | Beta-glucanase, CALCIUM ION | | Authors: | Fita, I, Planas, A, Calisto, B.M, Addington, T. | | Deposit date: | 2008-05-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Re-engineering specificity in 1,3-1,4-beta-glucanase to accept branched xyloglucan substrates
Proteins, 79, 2011
|
|
7BZL
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6LC5
 
 | | Crystal structure of barley exohydrolaseI W434F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LBB
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LBV
 
 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
8BJO
 
 | |
