1JPF
 
 | | Crystal Structure Of The LCMV Peptidic Epitope Gp276 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
1JPG
 
 | | Crystal Structure Of The LCMV Peptidic Epitope Np396 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
6SA6
 
 | | DARPin-Armadillo fusion A5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPin-Armadillo fusion A5 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Pluckthun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
4DBA
 
 | | Designed Armadillo repeat protein (YIIM3AII) | | Descriptor: | Designed Armadillo repeat protein, YIIM3AII, GLYCEROL | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DUI
 
 | | DARPIN D1 binding to tubulin beta chain (not in complex) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1 | | Authors: | Pecqueur, L, Duellberg, C, Dreier, B, Wang, Q, Jiang, C, Pluckthun, A, Surrey, T, Gigant, B, Knossow, M. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | An Anti-Tubulin Darpin Caps the Microtubule Plus-End
To be Published
|
|
4DB6
 
 | | Designed Armadillo repeat protein (YIIIM3AII) | | Descriptor: | Armadillo repeat protein | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB8
 
 | | Designed Armadillo-repeat Protein | | Descriptor: | Armadillo-repeat Protein, MAGNESIUM ION | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB9
 
 | | Designed Armadillo repeat protein (YIIIM3AIII) | | Descriptor: | Armadillo repeat protein, YIIIM3AIII | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
6S9N
 
 | | Designed Armadillo Repeat protein Lock2 fused to target peptide KRKRKAKLSF | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lock2_KRKRKAKLSF | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6S9L
 
 | | Designed Armadillo Repeat protein Lock1 bound to (KR)4KLSF target | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, KR4KLSF Lock1, LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG-LYS-LEU-SER-PHE | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6S9O
 
 | | Designed Armadillo Repeat protein internal Lock1 fused to target peptide KRKRKLKFKR | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, designed Armadillo repeat protein with internal Lock1 fused to target peptide KRKRKLKFKR | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6S9M
 
 | | Designed Armadillo Repeat protein Lock2 fused to target peptide KRKRKAKITW | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lock2_KRKRKAKITW, ... | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6S9P
 
 | | Designed Armadillo Repeat protein internal Lock2 fused to target peptide KRKAKITWKR | | Descriptor: | 1,2-ETHANEDIOL, internal Lock2 fused to target peptide KRKAKITWKR | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6SH9
 
 | | EngBF DARPin Fusion 4b D12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-alpha-N-acetylgalactosaminidase,DARPin 4b D12, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-08-06 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6S9T
 
 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 3 | | Descriptor: | Darpin 3, LIM domain-binding protein 1, TETRAETHYLENE GLYCOL | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S9S
 
 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 10 | | Descriptor: | Darpin 10, LIM domain-binding protein 1 | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FJ3
 
 | | High resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. | | Descriptor: | ACETIC ACID, CHLORIDE ION, OLEIC ACID, ... | | Authors: | Ehrenmann, J, Schoppe, J, Klenk, C, Rappas, M, Kummer, L, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FP7
 
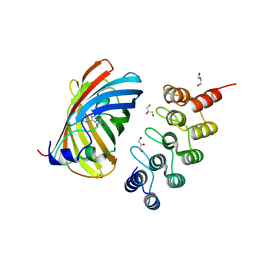 | | mTFP1/DARPin 1238_E11 complex in space group P6522 | | Descriptor: | DARPin1238_E11, GFP-like fluorescent chromoprotein cFP484, GLYCEROL | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP8
 
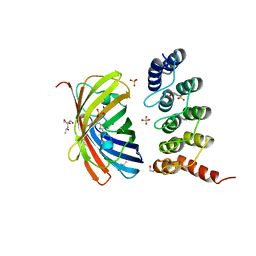 | | mTFP1/DARPin 1238_E11 complex in space group C2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DARPin1238_E11, ... | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPB
 
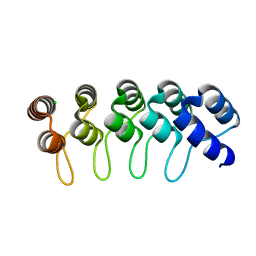 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group I4 | | Descriptor: | CHLORIDE ION, DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPA
 
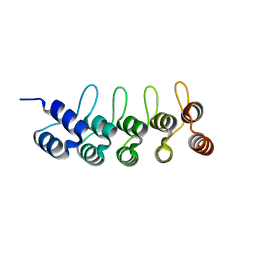 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6GCU
 
 | | MET receptor in complex with InlB internalin domain and DARPin A3A | | Descriptor: | DARPin A3A, Hepatocyte growth factor receptor, Internalin B | | Authors: | Meyer, T, Andres, F, Iamele, L, Gherardi, E, Pluckthun, A, Niemann, H.H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.001 Å) | | Cite: | Inhibition of the MET Kinase Activity and Cell Growth in MET-Addicted Cancer Cells by Bi-Paratopic Linking.
J.Mol.Biol., 431, 2019
|
|
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
1FG2
 
 | | CRYSTAL STRUCTURE OF THE LCMV PEPTIDIC EPITOPE GP33 IN COMPLEX WITH THE MURINE CLASS I MHC MOLECULE H-2DB | | Descriptor: | BETA-2 MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Tissot, A.C, Ciatto, C, Mittl, P.R.E, Gruetter, M.G, Plueckthun, A. | | Deposit date: | 2000-07-27 | | Release date: | 2000-10-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.754 Å) | | Cite: | Viral escape at the molecular level explained by quantitative T-cell receptor/peptide/MHC interactions and the crystal structure of a peptide/MHC complex.
J.Mol.Biol., 302, 2000
|
|
4F61
 
 | | Tubulin:Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gigant, B, Mignot, I, Knossow, M. | | Deposit date: | 2012-05-14 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Design and characterization of modular scaffolds for tubulin assembly.
J.Biol.Chem., 287, 2012
|
|
