2XUZ
 
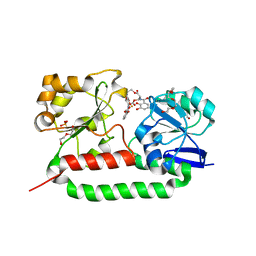 | | Crystal structure of the triscatecholate siderophore binding protein FeuA from Bacillus subtilis complexed with Ferri-Enterobactin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, IRON-UPTAKE SYSTEM-BINDING PROTEIN, ... | | Authors: | Peuckert, F, Miethke, M, Schwoerer, C.J, Albrecht, A.G, Oberthuer, M, Marahiel, M.A. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Siderophore Binding Protein Feua Shows Limited Promiscuity Toward Exogenous Triscatecholates
Chem.Biol., 18, 2011
|
|
2JUY
 
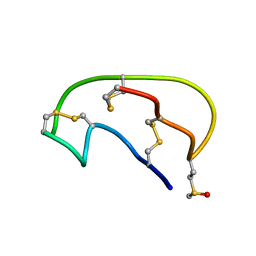 | | NMR ensemble of Neopetrosiamide A | | Descriptor: | Neopetrosiamide A | | Authors: | Austin, P, Williams, D.E, Heller, M, McIntosh, L.P, Andersen, R.J, Roberge, M, Roskelley, C.D. | | Deposit date: | 2007-09-05 | | Release date: | 2008-08-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR ensemble of Neopetrosiamide A
To be Published
|
|
3C58
 
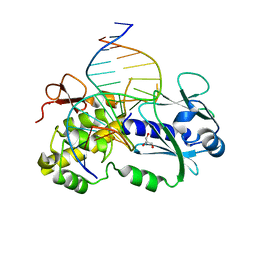 | | Crystal structure of a complex between the wild-type lactococcus lactis Fpg (MutM) and a N7-Benzyl-Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DCP*DTP*DTP*DTP*(SOS)P*DTP*DTP*DTP*DCP*DTP*DCP*DG)-3'), DNA (5'-D(*DGP*DCP*DGP*DAP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DGP*DA)-3'), DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Carell, T. | | Deposit date: | 2008-01-31 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode
Chem.Biol., 15, 2008
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QEC
 
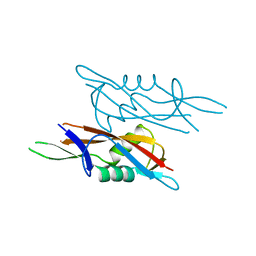 | | Crystal structure of SlpA - domain II, domain that is involved in the self-assembly of the S-layer from Lactobacillus amylovorus | | Descriptor: | S-layer | | Authors: | Eder, M, Dordic, A, Millan, C, Sagmeister, T, Uson, I, Pavkov-Keller, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QEH
 
 | | LTA-binding domain of SlpA, the S-layer protein from Lactobacillus amylovorus | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Pavkov-Keller, T. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFI
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain I (aa 31-182) | | Descriptor: | CALCIUM ION, SlpX | | Authors: | Sagmeister, T, Damisch, E, Millan, C, Uson, I, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFK
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II, Co-Crystallization with HgCl2, Mutation Ser316Cys (aa 194-362) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFG
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain III (aa 309-444) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, S-layer protein | | Authors: | Sagmeister, T, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFL
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain II (aa 199-308) | | Descriptor: | ACETATE ION, PHOSPHATE ION, S-layer protein | | Authors: | Sagmeister, T, Dordic, A, Eder, E, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLH
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 48-213) | | Descriptor: | PHOSPHATE ION, S-layer, SODIUM ION | | Authors: | Grininger, C, Sagmeister, T, Eder, E, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLE
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I (aa 32-198) | | Descriptor: | S-layer protein | | Authors: | Sagmeister, T, Eder, M, Vejzovic, D, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLD
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I, Co-crystallization with HgCl2, Mutation Ser146Cys, (aa 32-198) | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, S-layer protein | | Authors: | Sagmeister, T, Vejzovic, D, Eder, M, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8Q1O
 
 | | S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 32-209), important for Self-assembly | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Sagmeister, T, Grininger, C, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8AOL
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain III (aa 363-499) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sagmeister, T, Damisch, E, Eder, M, Dordic, A, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ALU
 
 | | Crystal structure of the teichoic acid binding domain of SlpA, S-layer protein from Lactobacillus acidophilus (aa. 314-444) | | Descriptor: | PHOSPHATE ION, S-layer protein | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8BT9
 
 | |
3ZOH
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOC
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOE
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOD
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOG
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOF
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
5OK6
 
 | | Ubiquitin specific protease 11 USP11 - peptide F complex | | Descriptor: | 1,2-ETHANEDIOL, ALA-GLU-GLY-GLU-PHE-TYR-LYS-LEU-LYS-ILE-ARG-THR-PRO-AAR, GLYCEROL, ... | | Authors: | Spiliotopoulos, A, Dreveny, I. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11.
J.Biol.Chem., 294, 2019
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
