3ZGA
 
 | | Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN 4 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZPJ
 
 | | Crystal structure of Ton1535 from Thermococcus onnurineus NA1 | | Descriptor: | TON_1535 | | Authors: | Jeong, J.H, kim, Y.G. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structure of the Hypothetical Protein Ton1535 from Thermococcus Onnurineus Na1 Reveals Unique Structural Properties by a Left-Handed Helical Turn in Normal Alpha-Solenoid Protein.
Proteins, 82, 2014
|
|
6L7Q
 
 | |
3ZG8
 
 | | Crystal Structure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZG7
 
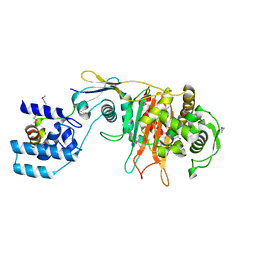 | |
3K1J
 
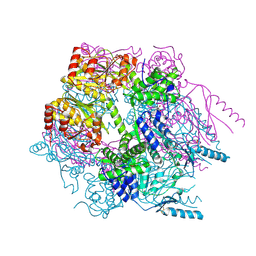 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
4EOG
 
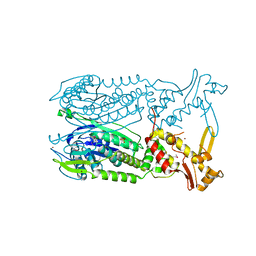 | | Crystal structure of Csx1 of Pyrococcus furiosus | | Descriptor: | Putative uncharacterized protein, SULFATE ION, ZINC ION | | Authors: | Kim, Y.K, Oh, B.H. | | Deposit date: | 2012-04-14 | | Release date: | 2013-01-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and nucleic acid-binding activity of the CRISPR-associated protein Csx1 of Pyrococcus furiosus.
Proteins, 81, 2013
|
|
7XI1
 
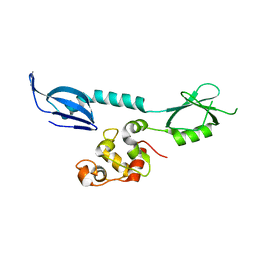 | | AcrIF 24 | | Descriptor: | anti-CRISPR protein AcrIF24 | | Authors: | Kim, G.E, Park, H.H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Molecular basis of dual anti-CRISPR and auto-regulatory functions of AcrIF24.
Nucleic Acids Res., 50, 2022
|
|
4UOY
 
 | | Crystal structure of YgjG in complex with Pyridoxal-5'-phosphate | | Descriptor: | FORMIC ACID, GLYCEROL, PUTRESCINE AMINOTRANSFERASE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2014-06-11 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structure of Putrescine Aminotransferase from Escherichia Coli Provides Insights Into the Substrate Specificity Among Class III Aminotransferases.
Plos One, 9, 2014
|
|
4UOX
 
 | |
5GL3
 
 | | Crystal structure of TON_0340 in complex with Mg | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL4
 
 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GKX
 
 | | Crystal structure of TON_0340, apo form | | Descriptor: | PHOSPHATE ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL2
 
 | | Crystal structure of TON_0340 in complex with Ca | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
2WWX
 
 | | Crystal structure of the SidM/DrrA(GEF/GDF domain)-Rab1(GTPase domain) complex | | Descriptor: | DRRA, RAS-RELATED PROTEIN RAB-1 | | Authors: | Suh, H.Y, Lee, D.W, Woo, J.S, Oh, B.H. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Dual Nucleotide Exchange and Gdi Displacement Activity of Sidm/Drra
Embo J., 29, 2010
|
|
7CHQ
 
 | | AcrIE2 | | Descriptor: | anti-CRISPR AcrIE2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A 1.3 angstrom high-resolution crystal structure of an anti-CRISPR protein, AcrI E2.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7D27
 
 | |
5XR6
 
 | | Crystal structure of RabA1a in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
5XPC
 
 | | Crystal Structure of Drep4 CIDE domain | | Descriptor: | DNAation factor-related protein 4, GLYCEROL | | Authors: | Park, H.H, Jeong, J.H. | | Deposit date: | 2017-06-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | CIDE domains form functionally important higher-order assemblies for DNA fragmentation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4I99
 
 | |
4I98
 
 | |
5AI1
 
 | | Crystal structure of ketosteroid isomerase containing Y32F, D40N, Y57F and Y119F mutations in the equilenin-bound form | | Descriptor: | EQUILENIN, KETOSTEROID ISOMERASE | | Authors: | Cha, H.J, Jeong, J.H, Kim, Y.G. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Contribution of a Low-Barrier Hydrogen Bond to Catalysis is not Significant in Ketosteroid Isomerase.
Mol.Cells, 38, 2015
|
|
