4PRK
 
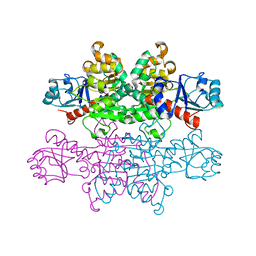 | |
3B1F
 
 | | Crystal structure of prephenate dehydrogenase from Streptococcus mutans | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative prephenate dehydrogenase | | Authors: | Ku, H.K, Do, N.H, Song, J.S, Choi, S, Shin, M.H, Kim, K.J, Lee, S.J. | | Deposit date: | 2011-07-02 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prephenate dehydrogenase from Streptococcus mutans.
Int.J.Biol.Macromol., 49, 2011
|
|
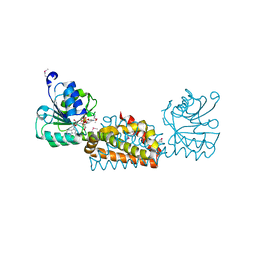
8GQM
 
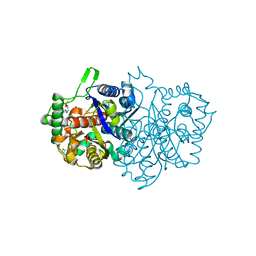 | |
8GQK
 
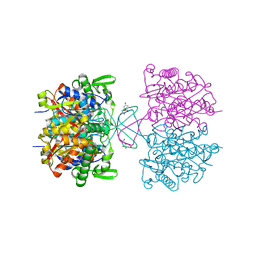 | |
8GQI
 
 | |
8GQJ
 
 | |
8GQF
 
 | |
8GQH
 
 | |
8GQN
 
 | |
8GQL
 
 | |
8GQG
 
 | |
8HGN
 
 | |
4PZE
 
 | |
4PZC
 
 | |
4PZD
 
 | |
7CW4
 
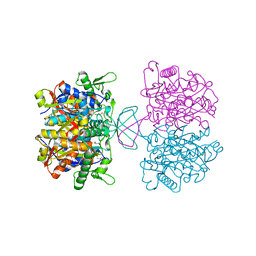 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CW5
 
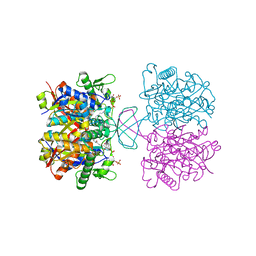 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
1G1B
 
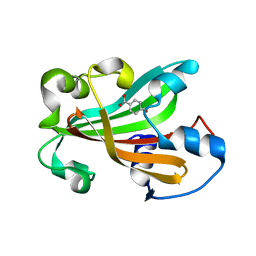 | | CHORISMATE LYASE (WILD-TYPE) WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Kim, K.J, Howard, A, Vilker, V.L. | | Deposit date: | 2000-10-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
5IWQ
 
 | |
5Z6U
 
 | |
5ELM
 
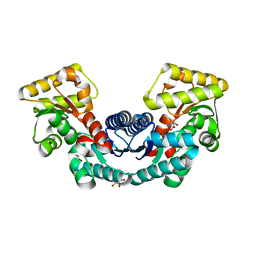 | | Crystal structure of L-aspartate/glutamate specific racemase in complex with L-glutamate | | Descriptor: | Asp/Glu_racemase family protein, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Ahn, J.W, Chang, J.H, Kim, K.J. | | Deposit date: | 2015-11-04 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for an atypical active site of an l-aspartate/glutamate-specific racemase from Escherichia coli
Febs Lett., 589, 2015
|
|
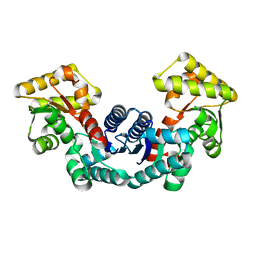
5ELL
 
 | |
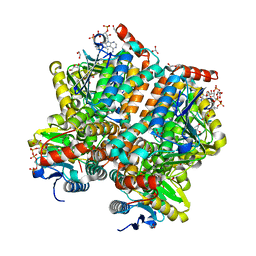
5ZAI
 
 | |
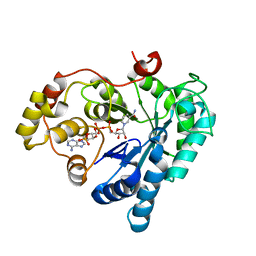
5Z6T
 
 | |
2CFR
 
 | |
