2CP0
 
 | | Solution structure of the 1st CAP-Gly domain in human CLIP-170-related protein CLIPR59 | | Descriptor: | CLIPR-59 protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 1st CAP-Gly domain in human CLIP-170-related protein CLIPR59
To be Published
|
|
2CP3
 
 | | Solution structure of the 2nd CAP-Gly domain in human CLIP-115/CYLN2 | | Descriptor: | CLIP-115 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 2nd CAP-Gly domain in human CLIP-115/CYLN2
To be Published
|
|
2COZ
 
 | | Solution structure of the CAP-Gly domain in human centrosome-associated protein CAP350 | | Descriptor: | centrosome-associated protein 350 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CAP-Gly domain in human centrosome-associated protein CAP350
To be Published
|
|
2COY
 
 | | Solution structure of the CAP-Gly domain in human Dynactin 1 | | Descriptor: | Dynactin-1 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CAP-Gly domain in human Dynactin 1
To be Published
|
|
2COW
 
 | | Solution structure of the CAP-Gly domain in human Kinesin-like protein KIF13B | | Descriptor: | Kinesin-like protein KIF13B | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CAP-Gly domain in human Kinesin-like protein KIF13B
To be Published
|
|
2CP7
 
 | | Solution structure of the 1st CAP-Gly domain in mouse CLIP-170/restin | | Descriptor: | Restin | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 1st CAP-Gly domain in mouse CLIP-170/restin
To be Published
|
|
2CP6
 
 | | Solution structure of the 2nd CAP-Gly domain in human CLIP-170/restin | | Descriptor: | Restin | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 2nd CAP-Gly domain in human CLIP-170/restin
To be Published
|
|
2CP2
 
 | | Solution structure of the 1st CAP-Gly domain in human CLIP-115/CYLN2 | | Descriptor: | CLIP-115 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 1st CAP-Gly domain in human CLIP-115/CYLN2
To be Published
|
|
2YUJ
 
 | |
2YUF
 
 | |
2YUG
 
 | |
2YUI
 
 | | Solution structure of the N-terminal domain in human cytokine-induced apoptosis inhibitor anamorsin | | Descriptor: | Anamorsin | | Authors: | Saito, K, Tomizawa, T, Tochio, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain in human cytokine-induced apoptosis inhibitor anamorsin
To be Published
|
|
2YUH
 
 | |
2YCD
 
 | | Structure of a novel Glutathione Transferase from Agrobacterium tumefaciens. | | Descriptor: | GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, S-(P-NITROBENZYL)GLUTATHIONE | | Authors: | Skopelitou, K, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2011-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Glutathione Transferase from Agrobacterium Tumefaciens Reveals a Novel Class of Bacterial Gst Superfamily.
Plos One, 7, 2012
|
|
3AGK
 
 | | Crystal structure of archaeal translation termination factor, aRF1 | | Descriptor: | Peptide chain release factor subunit 1 | | Authors: | Kobayashi, K, Kikuno, I, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Omnipotent role of archaeal elongation factor 1 alpha (EF1{alpha}) in translational elongation and termination, and quality control of protein synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2DCM
 
 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2DQ6
 
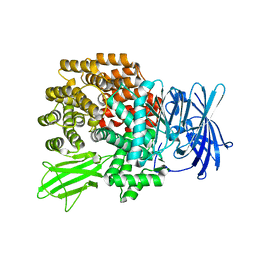 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2D5L
 
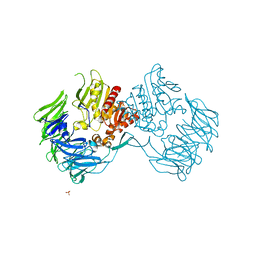 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2DQM
 
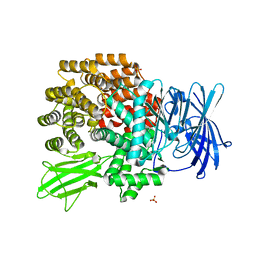 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2CZH
 
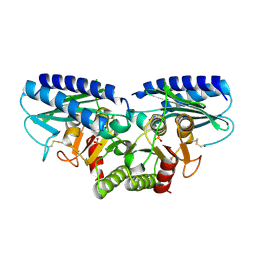 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) with phosphate ion (orthorhombic form) | | Descriptor: | Inositol monophosphatase 2, PHOSPHATE ION | | Authors: | Ito, K, Arai, R, Kamo-Uchikubo, T, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
1WEG
 
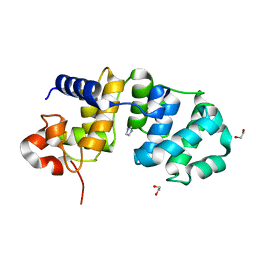 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
1WEF
 
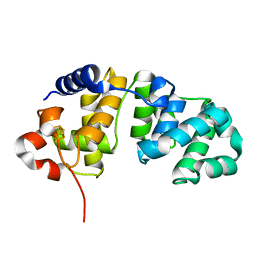 | |
1WEI
 
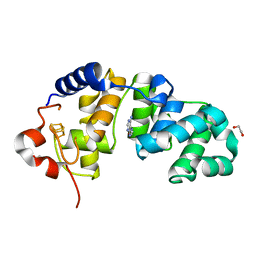 | | Catalytic Domain Of Muty From Escherichia Coli K20A Mutant Complexed To Adenine | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, ADENINE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
4KS6
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 166-174 stretch | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-17 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
4KTX
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 167-174 stretch | | Descriptor: | Botulinum neurotoxin A light chain, GLYCEROL, Peptide inhibitor MPT-DPP-ARG-G-LEU-NH2, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-21 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
