1W31
 
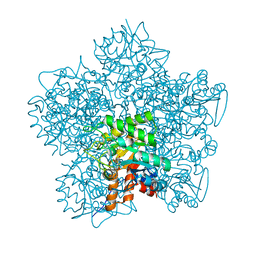 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | | Descriptor: | 5-HYDROXYLAEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Beaven, G.D.E, Gill, R, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2004-07-11 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UW7
 
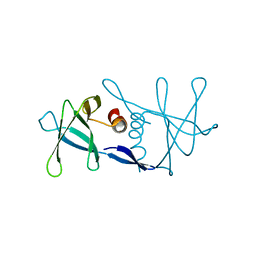 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
1QJV
 
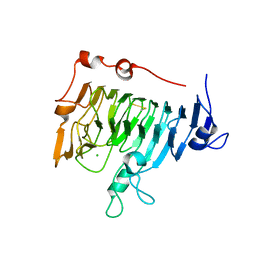 | | Pectin methylesterase PemA from Erwinia chrysanthemi | | Descriptor: | CHLORIDE ION, PECTIN METHYLESTERASE | | Authors: | Jenkins, J, Mayans, O, Smith, D, Worboys, K, Pickersgill, R. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site
J.Mol.Biol., 305, 2001
|
|
1XBU
 
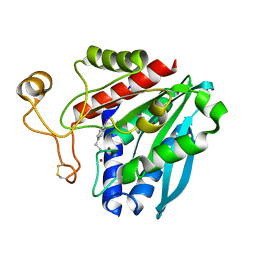 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
1W1Z
 
 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
1W6S
 
 | | The high resolution structure of methanol dehydrogenase from methylobacterium extorquens | | Descriptor: | CALCIUM ION, GLYCEROL, METHANOL DEHYDROGENASE SUBUNIT 1, ... | | Authors: | Williams, P.A, Coates, L, Mohammed, F, Gill, R, Erskine, P.T, Wood, S.P, Anthony, C, Cooper, J.B. | | Deposit date: | 2004-08-23 | | Release date: | 2004-12-21 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Atomic Resolution Structure of Methanol Dehydrogenase from Methylobacterium Extorquens
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X87
 
 | | 2.4A X-ray structure of Urocanase protein complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Urocanase protein | | Authors: | Tereshko, V, Zaborske, J, Gilbreth, R, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A X-ray structure of Urocanase protein complexed with NAD
To be Published
|
|
1ZC8
 
 | | Coordinates of tmRNA, SmpB, EF-Tu and h44 fitted into Cryo-EM map of the 70S ribosome and tmRNA complex | | Descriptor: | Elongation factor Tu, H2 16S rRNA, H2b d mRNA, ... | | Authors: | Valle, M, Gillet, R, Kaur, S, Henne, A, Ramakrishnan, V, Frank, J. | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Visualizing tmRNA Entry into a Stalled Ribosome
Science, 300, 2003
|
|
1H4J
 
 | | Methylobacterium extorquens methanol dehydrogenase D303E mutant | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 1, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Mohammed, F, Gill, R, Thompson, D, Cooper, J.B, Wood, S.P, Afolabi, P.R, Anthony, C. | | Deposit date: | 2001-05-11 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Directed Mutagenesis and X-Ray Crystallography of the Pqq-Containing Quinoprotein Methanol Dehydrogenase and its Electron Acceptor, Cytochrome C(L)(,)
Biochemistry, 40, 2001
|
|
1QN2
 
 | | cytochrome cH from Methylobacterium extorquens | | Descriptor: | CYTOCHROME CH, HEME C | | Authors: | Read, J, Gill, R, Dales, S.L, Cooper, J.B, Wood, S.P, Anthony, C. | | Deposit date: | 1999-10-13 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Molecular Structure of an Unusual Cytochrome C2 Determined at 2.0A; the Cytochrome cH from Methylobacterium Extorquens
Protein Sci., 8, 1999
|
|
1K3W
 
 | | Crystal structure of a trapped reaction intermediate of the DNA Repair Enzyme Endonuclease VIII with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1TKJ
 
 | | Streptomyces griseus aminopeptidase complexed with D-Methionine | | Descriptor: | Aminopeptidase, CALCIUM ION, D-METHIONINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
1K3X
 
 | | Crystal structure of a trapped reaction intermediate of the DNA repair enzyme Endonuclease VIII with Brominated-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1TF9
 
 | | Streptomyces griseus aminopeptidase complexed with P-Iodo-L-Phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, IODO-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TKH
 
 | | Streptomyces griseus aminopeptidase complexed with D-Phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
1TF8
 
 | | Streptomyces griseus aminopeptidase complexed with L-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TKF
 
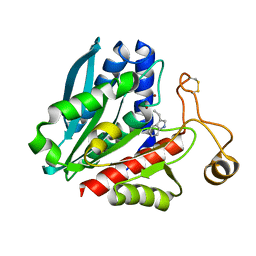 | | Streptomyces griseus aminopeptidase complexed with D-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, D-TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
5OT7
 
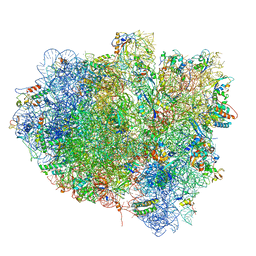 | | Elongation factor G-ribosome complex captures in the absence of inhibitors. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mace, K, Giudice, E, Chat, S, Gillet, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of an elongation factor G-ribosome complex captured in the absence of inhibitors.
Nucleic Acids Res., 46, 2018
|
|
2K4G
 
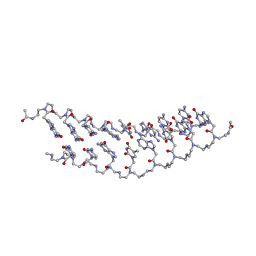 | | Solution Structure of a Peptide Nucleic Acid Duplex, 10 structures | | Descriptor: | METHYLAMINE, PNA (N'-(*(GPN)*(GPN)*(CPN)*(APN)*(TPN)*(GPN)*(CPN)*(CPN))-C') | | Authors: | He, W, Hatcher, E, Balaeff, A, Beratan, D, Gil, R, Madrid, M, Achim, C. | | Deposit date: | 2008-06-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a peptide nucleic acid duplex from NMR data: features and limitations.
J.Am.Chem.Soc., 130, 2008
|
|
4NEU
 
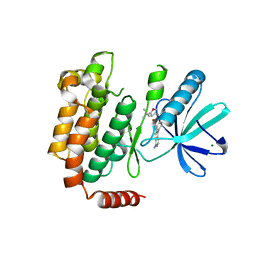 | | X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor | | Descriptor: | 1-[4-(1-aminoisoquinolin-5-yl)phenyl]-3-(5-tert-butyl-1,2-oxazol-3-yl)urea, MAGNESIUM ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Nolte, R.T, Ward, P, kahler, K.M, Campobasso, N. | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis.
ACS Med Chem Lett, 4, 2013
|
|
5HX6
 
 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5HNR
 
 | | The X-ray structure of octameric human native 5-aminolaevulinic acid dehydratase. | | Descriptor: | DELTA-AMINO VALERIC ACID, Delta-aminolevulinic acid dehydratase, SULFATE ION, ... | | Authors: | Mills-Davies, N.L, Thompson, D, Shoolingin-Jordan, P.M, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-01-18 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6YOA
 
 | | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family | | Descriptor: | Major pollen allergen Lig v 1, NICKEL (II) ION | | Authors: | Robledo-Retana, T, Bradley-Clark, J, Croll, T, Rose, R, Stagg, A, Villalba, M, Pickersgill, R. | | Deposit date: | 2020-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family.
Allergy, 75, 2020
|
|
1LES
 
 | | LENTIL LECTIN COMPLEXED WITH SUCROSE | | Descriptor: | CALCIUM ION, LENTIL LECTIN, MANGANESE (II) ION, ... | | Authors: | Hamelryck, T, Loris, R. | | Deposit date: | 1995-08-23 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NMR, molecular modeling, and crystallographic studies of lentil lectin-sucrose interaction.
J.Biol.Chem., 270, 1995
|
|
1LEN
 
 | | REFINEMENT OF TWO CRYSTAL FORMS OF LENTIL LECTIN AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Van Overberge, D, Loris, R, Wyns, L. | | Deposit date: | 1993-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
