3MAH
 
 | | A putative c-terminal regulatory domain of aspartate kinase from porphyromonas gingivalis w83. | | Descriptor: | Aspartokinase, SULFATE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Moy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Putative C-Terminal Regulatory Domain of Aspartate Kinase from Porphyromonas Gingivalis W83.
To be Published
|
|
3MVN
 
 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
3NE7
 
 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
4HTF
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase from Escherichia coli in complex with S-adenosylmethionine. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase from Escherichia coli in complex with S-adenosylmethionine.
To be Published
|
|
3NQR
 
 | | A putative CBS domain-containing protein from Salmonella typhimurium LT2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein corC | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Cui, H, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A putative CBS domain-containing protein from Salmonella typhimurium LT2.
To be Published
|
|
4HVN
 
 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 in complex with Trimethylamine. | | Descriptor: | N,N-dimethylmethanamine, hypothetical protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
3OET
 
 | | D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Edwards, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-13 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD
To be Published
|
|
3OOW
 
 | | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4.
To be Published
|
|
3OPQ
 
 | | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure. | | Descriptor: | CHLORIDE ION, FORMIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.
To be Published
|
|
3OXP
 
 | | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution | | Descriptor: | GLYCEROL, Phosphotransferase enzyme II, A component, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-21 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution
To be Published
|
|
4ICH
 
 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
3PEA
 
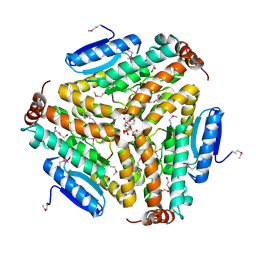 | | Crystal structure of enoyl-CoA hydratase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | ACETATE ION, CITRATE ANION, Enoyl-CoA hydratase/isomerase family protein, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
4ISC
 
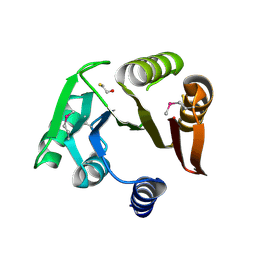 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4JJX
 
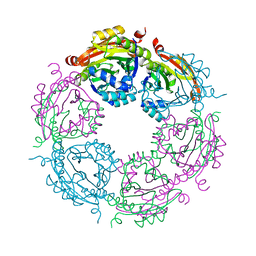 | | Dodecameric structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae O1 biovar eltor | | Descriptor: | Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-08 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4JLY
 
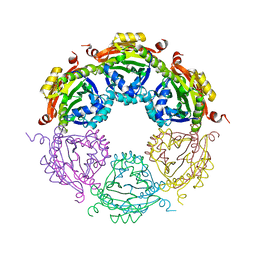 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
3PJ9
 
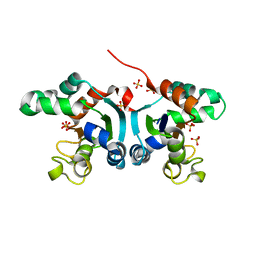 | | Crystal structure of a Nucleoside Diphosphate Kinase from Campylobacter jejuni | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a Nucleoside Diphosphate Kinase from Campylobacter jejuni
To be Published
|
|
4JNN
 
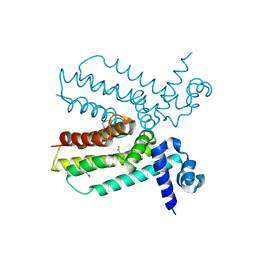 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine | | Descriptor: | BENZAMIDINE, BETA-MERCAPTOETHANOL, Transcriptional regulator | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-15 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine
To be Published
|
|
4JG2
 
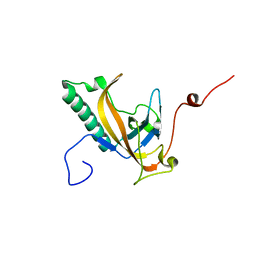 | | Structure of phage-related protein from Bacillus cereus ATCC 10987 | | Descriptor: | Phage-related protein | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of phage-related protein from Bacillus cereus ATCC 10987
To be Published
|
|
3P0R
 
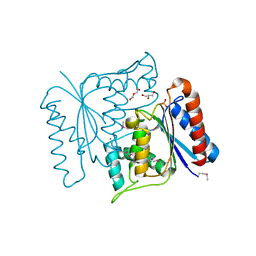 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
4KMR
 
 | | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Transcriptional regulator, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542.
To be Published
|
|
3QFG
 
 | | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Halavaty, A, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
4JBF
 
 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
3OUT
 
 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
3P52
 
 | | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NITRATE ION | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion
To be Published
|
|
3R2I
 
 | | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Exotoxin 5 | | Authors: | Filippova, E.V, Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
